Figures & data
Figure 2. The mitochondrial genome map of Megalurothrips usitatus. Genes are shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. GC and AT contents across the mitochondrial genome is shown with dark and light shading, respectively, inside the inner circle.
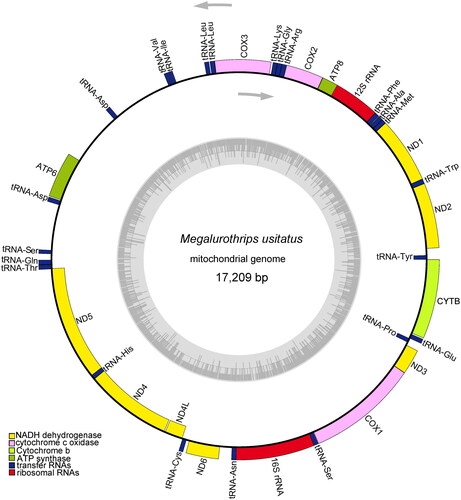
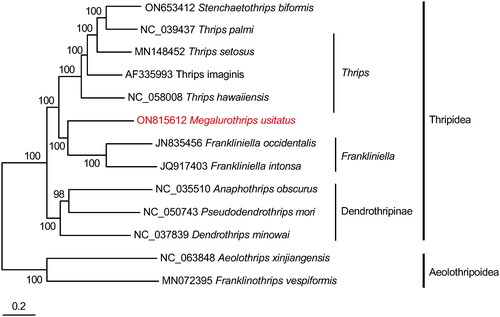
Figure 3. Phylogenetic tree of 13 insect species, including Megalurothrips usitatus based on the nucleotide dataset of the 13 mitochondrial protein-coding genes and ribosomal genes. ‘GTR + I + G’ was used as the best-fit nucleotide substitution model. Bayesian Inference phylogenies were inferred using Markov Chain Monte Carlo (one cold and three hot chains) chains of 2,000,000 with 25% burn-in, and sampling was done every 100 generation. The posterior probabilities are indicated above the nodes. The GenBank accession no. of sequences used in the study are stenchaetothrips biformis ON653412 (Hu et al. Citation2023); Thrips palmi NC_039437 (Chakraborty et al. Citation2018); Thrips setosus MN148452; Thrips imaginis AF335993 (Shao and Barker Citation2003); Thrips hawaiiensis NC_058008; Megalurothrips usitatus ON815612 (this study); Frankliniella occidentalis JN835456 (Yan et al. Citation2012); Frankliniella intonsa JQ917403 (Yan et al. Citation2014); dendrothrips minowai NC_037839; anaphothrips obscurus NC_035510 (Liu et al. Citation2017); pseudodendrothrips mori NC_050743; Aeolothrips xinjiangensis NC_063848; Franklinothrips vespiformis MN072395 (Tyagi et al. Citation2020).
Supplemental Material
Download TIFF Image (5 MB)Supplemental Material
Download MS Word (163.8 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON815612. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA935568, SRP423475 and SAMN33321779, respectively.

