Figures & data
Figure 1. Photographs of M. elongata (the photograph were taken by prof. Haimei Chen). the image was captured at the Horticulture and Landscape Architecture College of Southwest University, Chongqing, China. The geospatial reference for this location is N29.842889, E106.394527. This visual representation demonstrates the distinctive features of the compact cactus, which is native to Mexico and highly appreciated for its ease of propagation in horticultural practice.

Figure 2. The chloroplast genome of Mammillaria elongata visualized by CPGView. The outer circle depicts the locations of protein-coding genes, tRNA genes, and rRNA genes along the 110,981 bp circular chloroplast chromosome. Genes are color-coded by function: photosystem I (orange), photosystem II (red), cytochrome b/f complex (purple), ATP synthase (blue), rubisco large subunit (green), RNA polymerase (teal), ribosomal proteins (small subunit in pink, large subunit in grey), other (light blue). The inner circle indicates the genome position in kilobase pairs.
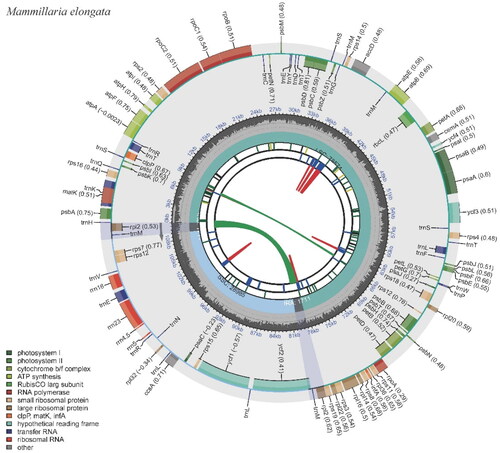
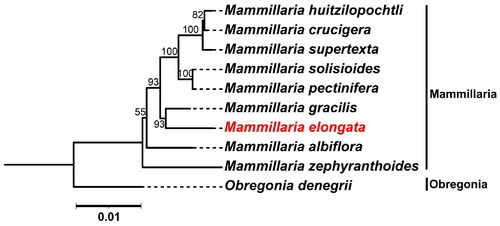
Figure 3. Maximum Likelihood (ML) phylogenetic tree Illustrating the relationship between M. elongata and eight other species within the Mammillaria genus based on chloroplast genome sequences. Obregonia species are utilized as outgroup references. M. elongata (MW553058.1) is distinctively marked in red. The sequences incorporated in the tree are as follows: M. huitzilopochtli (MN517612.1) (Solórzano et al., Citation2019), M. supertexta (MN508963.1) (Solórzano et al., Citation2019), M. solisioides (MN518341.1) (Solórzano et al., Citation2019), M. pectinifera (MN519716.1) (Solórzano et al., Citation2019), M. gracilis (MW553059.1) (Yu et al., Citation2023), M. albiflora (MN517610.1) (Solórzano et al., Citation2019), M. zephyranthoides (MN517611.1) (Solórzano et al., Citation2019), and O. denegrii (MW553062.1) (Yu et al., Citation2023). the scale bar represents a genetic distance of 0.1 substitutions per site.
Supplemental Material
Download MS Word (205.6 KB)Data availability statement
The genome sequence data supporting this study’s findings have been deposited in the NCBI GenBank and can be accessed at https://www.ncbi.nlm.nih.gov/ with the accession number MW553058.1. Corresponding BioProject, Sequence Read Archive (SRA), and BioSample entries are available under the identifiers PRJNA995908, SRR25317666, and SAMN36509928, respectively.
