Figures & data
Figure 1. Phyla nodiflora plant used in this study. A. Collected sample, B. Habitat, and C. Digital herbarium (MH178191). The features that differentiate P. nodiflora from the other species of this genus are obovate leaves with sub-obtuse apex, pinnate venation prominent abaxially, serrate leaf margin, spherical-shaped fruit and rooting at the nodes (O'Leary and Múlgura Citation2012). These images were taken by authors (Ray Sharmishtha and Tanuja). the plant sample was collected from Guduvanchery, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°51′16.1″N 80°03′49.4″E) on 9 September 2022.

Figure 2. Circular map of the chloroplast genome of Phyla nodiflora. From the center going outward, the first circle shows the distribution of the repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle displays the tandem repeats marked with short bars. The third circle shows the LSC, SSC, IRa, and IRb regions. The fourth circle shows the percentage of GC content. The next circle shows the genes having different colors based on the functional groups. The functional classification is shown at the bottom left. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counter-clockwise direction.
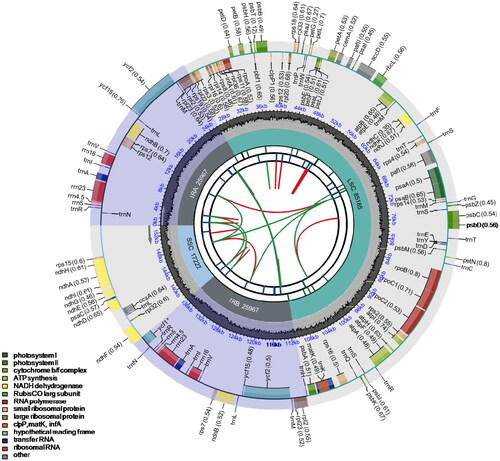
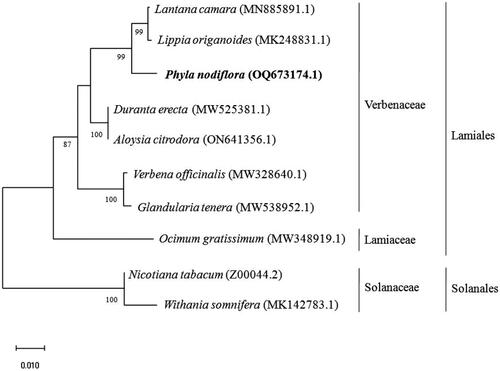
Figure 3. Phylogenetic tree constructed by maximum likelihood analysis based on complete chloroplast genome sequences, including the chloroplast genome of P. nodiflora (OQ673174). The bootstrap support values are shown on the nodes. The chloroplast genome sequences used for the phylogenetic analysis were Ocimum gratissimum MW348919.1 (Balaji et al. Citation2021), Duranta erecta MW525381.1 (Song et al. Citation2021), Aloysia citrodora ON641356.1, Verbena officinalis MW328640.1 (Yue et al. Citation2021), Phyla nodiflora OQ673174.1, Lanata camera MN885891.1, Lippia origanoides MK248831.1 (Sarzi et al. Citation2019), Nicotiana tabacum Z00044.2 (Shinozaki et al. Citation1986), and Withania somnifera MK142783.1 (Mehmood et al. Citation2020).
Supplemental Material
Download TIFF Image (74.2 MB)Supplemental Material
Download TIFF Image (2.9 MB)Supplemental Material
Download TIFF Image (9.2 MB)Data availability statement
The genome sequence data supporting the findings of this study are available in NCBI under the accession number OQ673174 (https://www.ncbi.nlm.nih.gov/nuccore/OQ673174.1). The associated next-generation sequencing data files are available from the BioProject, Bio-Sample, and SRA submission under the accession numbers PRJNA945246, SAMN33776799, and SRR23875832, respectively.
