Figures & data
Figure 1. The species reference image for E. alashanicus. Morphological characteristics of whole plant (A), root (B), ligule (C), glumes (D), spikelet (E), and inflorescence (F) of E. alashanicus (photos taken by author Feng Jin in Helan Mountain, Inner Mongolia, China).
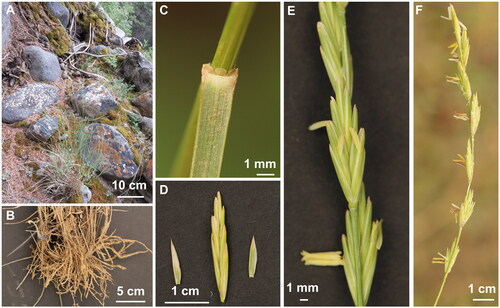
Figure 2. Schematic circular map of overall features of E. alashanicus chloroplast genome. Graphic showing features of its plastome was generated using CPGview. The map contains six tracks. From the inner circle, the first track depicts the dispersed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The second track shows the long tandem repeats as short blue bars. The third track displays the short tandem repeats or microsatellite sequences as short bars with different colors. The fourth track depicts the sizes of the inverted repeats (IRa and IRb), small single-copy (SSC), and large single-copy (LSC). The fifth track plots the distribution of GC contents along the plastome. The sixth track displays the genes belonging to different functional groups with different colored boxes. The outer and inner genes are transcribed in the clockwise and counterclockwise directions, respectively.
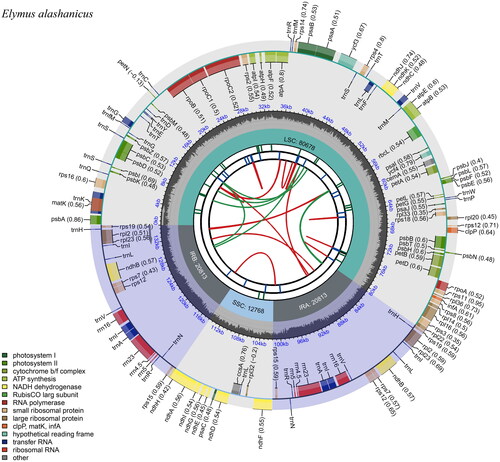
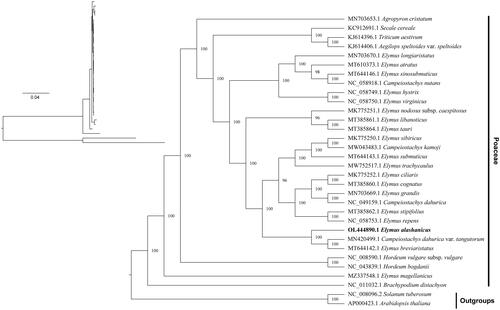
Figure 3. Maximum-likelihood and Bayesian inference phylogenetic trees of 30 Poaceae species with Arabidopsis thaliana and Solanum tuberosum as outgroups. The main phylogenetic tree was presented based on the BI tree, because of highly similarity among the sequences, showed lower branch support of the ML evolutionary tree. Species names are displayed with indicate lines in the right side, and the numbers above branches indicate supportive values of BI phylogenetic trees. GenBank accession numbers of the following sequences were used to construct the phylogenetic tree: Agropyron cristatum MN703653.1 (Chen et al. Citation2018), Secale cereale KC912691.1 (Li et al. Citation2015), Triticum aestivum KJ614396.1 (Han et al. Citation2021), Aegilops speltoides var. speltoides KJ614406.1 (Chen et al. Citation2018), Elymus longiaristatus MN703670.1 (Hu et al. Citation2015), Elymus atratus MT610373.1 (Gao et al. Citation2014), Elymus sinosubmuticus MT644146.1 (Tan et al. Citation2022), Campeiostachys nutans NC_058918.1 (Dong et al. Citation2015), Elymus hystrix NC_058749.1 (Dong et al. Citation2015), Elymus virginicus NC_058750.1 (Dong et al. Citation2015), Elymus nodosus subsp. caespitosus MK775251.1 (Chen et al. Citation2020), Elymus libanoticus MT385861.1 (Xia and Liu Citation2020), Elymus tauri MT385864.1 (Liu et al. Citation2021), Elymus sibiricus MK775250.1 (Chen et al. Citation2022), Campeiostachys kamoji MW043483.1 (Liu et al. Citation2021), Elymus submuticus MT644143.1 (Y. NI 2011), Elymus trachycaulus MW752517.1 (Wu et al. Citation2016), Elymus ciliaris MK775252.1 (Hu et al. Citation2013), Elymus cognatus MT385860.1 (Hu et al. Citation2015), Elymus grandis MN703669.1 (Yu et al. Citation2010), Campeiostachys dahurica NC_049159.1 (Tan et al. Citation2021), Elymus stipifolius MT385862.1 (Yang et al. Citation2017), Elymus repens NC_058753.1 (Mason-Gamer Citation2008), Campeiostachys dahurica var. tangutorum MN420499.1 (Yang et al. Citation2015; Jing et al. Citation2019), Elymus breviaristatus MT644142.1 (Tan et al. Citation2022), Hordeum vulgare subsp. vulgare NC_008590.1 (Zhelyazkova et al. Citation2012), Hordeum bogdanii NC_043839.1 (Cui et al. Citation2021), Elymus magellanicus MZ337548.1 (Wu et al. Citation2022), Brachypodium distachyon NC_011032.1 (Sancho et al. Citation2018), Solanum tuberosum NC_008096.2 (Occhialini et al. Citation2020), and Arabidopsis thaliana AP000423.1 (Provan Citation2000).
Supplemental Material
Download TIFF Image (333.9 KB)Supplemental Material
Download TIFF Image (800.4 KB)Supplemental Material
Download TIFF Image (757.1 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OL444890.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA778469, SRR16913047, and SAMN22960735, respectively.
