Figures & data
Figure 1. Photograph of the Cyphocaris challengeri specimen sequenced in this study. Scale bar 1 mm. Photo taken by C. Rabinowitz.
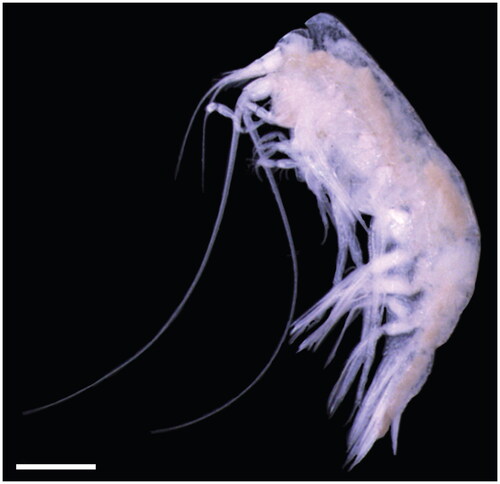
Figure 2. Circular mitogenome map of Cyphocaris challengeri. All annotated genes are indicated with their direction. Inner tract shows GC content with an 80 bp sliding window; high (51% GC) and low (5% GC) values are bounded by the gray rings.
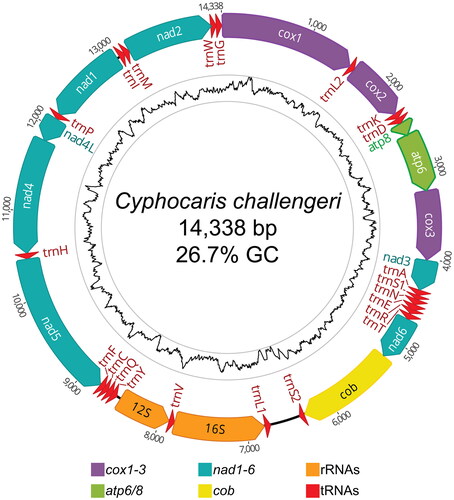
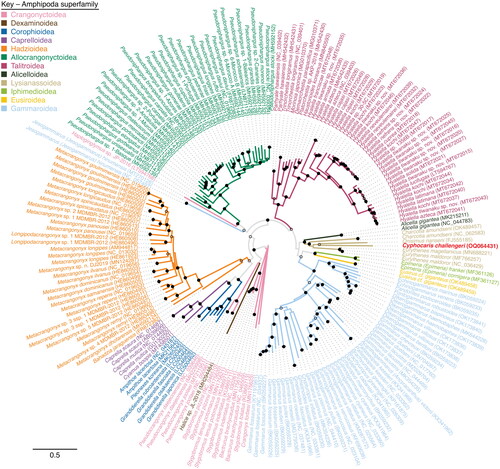
Figure 3. Maximum likelihood phylogenetic tree of a concatenated alignment of all thirteen protein encoding genes available from Amphipoda mitogenomes. Cyphocaris challengeri label is highlighted in red. Branches are colored by superfamily. Bootstrap values are indicated at nodes with a solid dot (≥75% support), open dot (≥50%, <75% support), or no dot (<50% support).
Supplemental Material
Download TIFF Image (4 MB)Supplemental Material
Download MS Excel (17.5 KB)Data availability statement
The complete mitochondrial genome of Cyphocaris challengeri is available on NCBI under accession number OQ064431. The complete rRNA-ITS region of two haplotypes (A/B) is available under accession number OQ093270 and OQ093271, respectively. The raw sequencing data from the C. challengeri genome is available through NCBI’s SRA, under BioProject, SRA, and BioSample accession numbers PRJNA913968, SRR22972441, and SAMN32316678, respectively.
