Figures & data
Figure 1. Swertia divaricate. Swertia divaricate is a perennial herb, has divaricate inflorescences and blackish purple corolla. Photographs by Pengcheng Fu in Gongshan, Yunnan Province, China.

Figure 2. Schematic map of the chloroplast genome of Swertia divaricata generated using CPGview. From center outwards, first circle shows distributed repeats connected with red (forward direction) and green (reverse direction) arcs. Second circle shows tandem repeats marked with short bars. Third circle shows microsatellite sequences as short bars. Forth circle shows sizes of LSC: long single copy, SSC: short single copy regions and inverted repeat (IRa and IRb) region. Fifth circle shows GC contents along the genome. Sixth circle shows genes marked with different colors according to their functional groups.
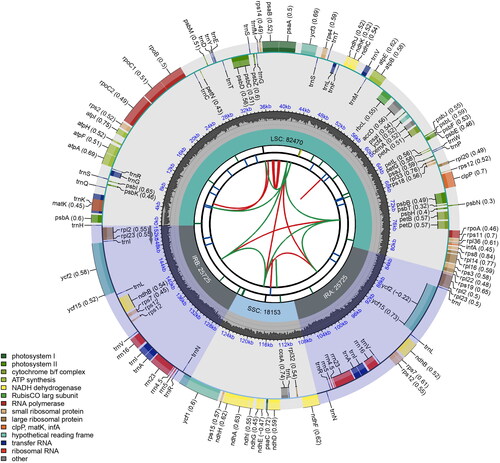
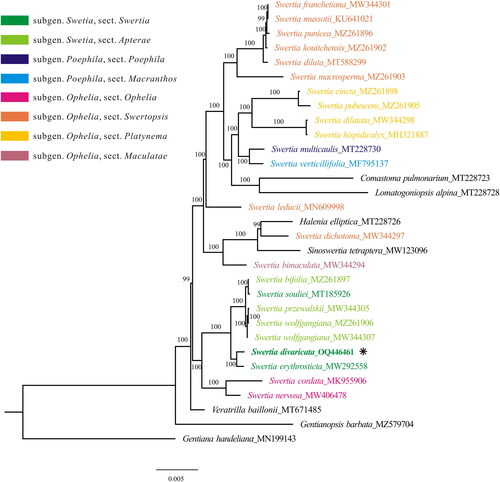
Figure 3. The maximum likelihood tree of Swertia, constructed using protein-coding genes from the plastome. Numbers above/below branches present bootstrap support. Species shown in bold is the newly sequenced in this study. The newly sequenced species was marked with an asterisk (*). The sectional classification of Swertia were marked with variable colors according to the treatment in Ho and Liu (Citation2015). the following sequences were used: S. franchetiana MW344301 (Xu et al. Citation2021), S. mussotii KU641021 (Xiang et al. Citation2016), S. punicea MZ261896 (Cao et al. Citation2022), S. kouitchensis MZ261902 (Cao et al. Citation2022), S. diluta MT588299 (Yang et al. Citation2020a), S. macrosperma MZ261903 (Cao et al. Citation2022), S. cincta MZ261898 (Cao et al. Citation2022), S. pubescens MZ261905 (Cao et al. Citation2022), S. dilatata MW344298 (Xu et al. Citation2021), S. hispidicalyx MH321887 (unpublished), S. multicaulis MT228730 (Zhang et al. Citation2020a, Citation2020b), S. verticillifolia MF795137 (Zhang et al. Citation2020a, Citation2020b), S. leducii MN609998 (unpublished), S. dichotoma MW344297 (Xu et al. Citation2021), S. bimaculata MW344294 (Xu et al. Citation2021), S. bifolia MZ261897 (Cao et al. Citation2022), S. souliei MT185926 (Bi et al. Citation2020), S. przewalskii MW344305 (Xu et al. Citation2021), S. wolfgangiana MZ261906 (Cao et al. Citation2022) MW344307 (Xu et al. Citation2021), S. erythrosticta MW292558 (unpublished), S. cordata MK955906 (Huang et al. Citation2019), S. nervosa MW406478 (Xu et al. Citation2021), Comastoma pulmonarium MT228723 (Zhang et al. Citation2020a, Citation2020b), Lomatogoniopsis alpina MT228728 (Zhang et al. Citation2020a, Citation2020b), Halenia elliptica MT228726 (Zhang et al. Citation2020a, Citation2020b), Sinoswertia tetraptera MW123096 (Yang et al. Citation2020b), veratrilla baillonii MT671485 (unpublished), Gentianopsis barbata MZ579704 (Feng et al. Citation2022), Gentiana handeliana MN199143 (Fu et al. Citation2022).
Supplemental Material
Download JPEG Image (417.7 KB)Supplemental Material
Download JPEG Image (1.2 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. OQ446461. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA935068, SAMN33296155, and SRR23490124, respectively.
