Figures & data
Figure 1. Species reference image of Geum longifolium in this study. (A) whole plant; (B) basal leaf; (C) flower. Species images were taken by the corresponding author Qin-Qin Li in Qilian county, Qinghai province, China.
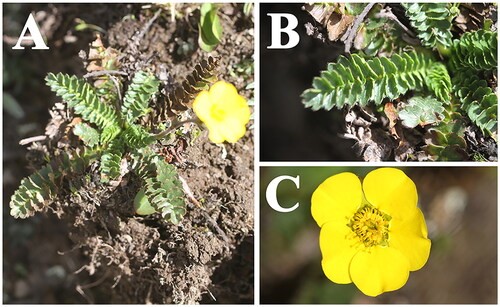
Figure 2. Genome map of G. longifolium chloroplast genome drawn by Chloroplast Genome Viewer (CPGView, http://www.1kmpg.cn/cpgview). The genome map includes six tracks. From the inward to outward, the first track shows the dispersed repeats which consist of direct repeats and palindromic repeats, connected with red and green arcs. The second track shows the long tandem repeats (blue bars). The third track shows the short tandem repeats or microsatellite sequences as short bars. The fourth track shows the large single copy (LSC), the small single copy (SSC), and inverted repeat (IRa and IRb) regions. The fifth track shows the GC contents along the chloroplast genome. The outermost track shows the genes which are color-coded based on their functional classification. The inner genes are transcribed clockwise, and the outer genes are transcribed anticlockwise.
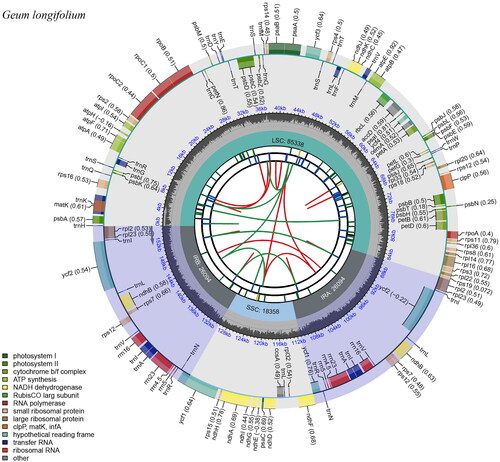
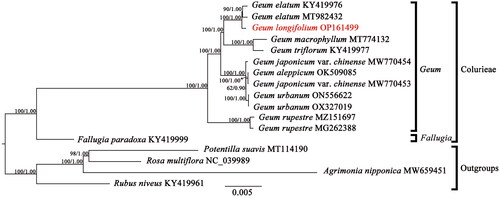
Figure 3. The Maximum-likelihood (ML) phylogenetic tree reconstructed based on 13 cp genome sequences from Colurieae plus four other Rosoideae species as outgroups. Values along branch represent ML bootstrap percentages, and Bayesian posterior probabilities respectively. The following sequences were used: Geum elatum KY419976 (Zhang et al. Citation2017), Geum elatum MT982432, Geum longifolium OP161499, Geum macrophyllum MT774132 (Li and Wen Citation2021), Geum triflorum KY419977 (Zhang et al. Citation2017), Geum japonicum var. chinense MW770454, Geum japonicum var. chinense MW770453, Geum aleppicum OK509085 (Zhang et al. Citation2022), Geum urbanum ON556622, Geum urbanum OX327019, Geum rupestre MZ151697, Geum rupestre MG262388 (Duan et al. Citation2018), fallugia paradoxa KY419999 (Zhang et al. Citation2017), Potentilla suavis MT114190 (Li et al. Citation2020), Rosa multiflora NC_039989, Agrimonia nipponica MW659451, and Rubus niveus KY419961 (Zhang et al. Citation2017).
Supplemental Material
Download JPEG Image (2.8 MB)Supplemental Material
Download JPEG Image (1.8 MB)Supplemental Material
Download MS Word (1.2 MB)Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OP161499. The associated BioProject, SRA, and Bio-Sample accession numbers are PRJNA866517, SRR21050029, and SAMN30167531, respectively.
