Figures & data
Figure 1. Photograph of Torenia violacea (the photograph was taken by prof. Xiaoyan Xiang in Mt. Huangshan, China). The foliage of T. violacea exhibits ovate or narrowly ovate shapes, characterized by villous, shallowly serrate margin, acuminate apex. The flowers are in terminal fascicles or solitary in leaf axils, rarely in racemes. Furthermore, the calyx of T. violacea displays purple-red and the corollas pale yellow or white.

Figure 2. Schematic map of overall features of the chloroplast genome of Torenia violacea. The boxes of different sizes and colors on the outermost circle represent genes and their lengths. The inner and outer boxes of the outermost circle represent genes transcribed clockwise and counter-clockwise. The gray area in the middle circle represents the changes in GC content at different positions, and the regions and lengths represented by the tetrad structure (LSC, SSC, IRa, and IRb) are drawn in different colors on the inner circle.
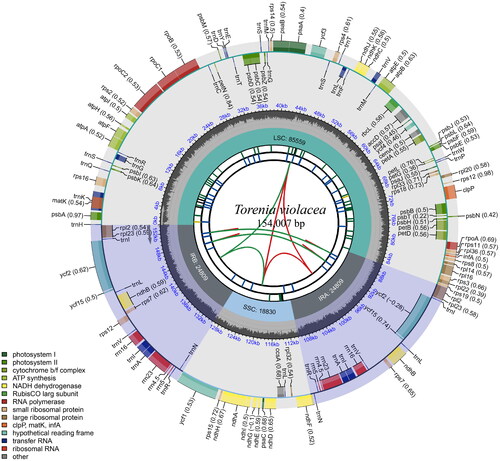
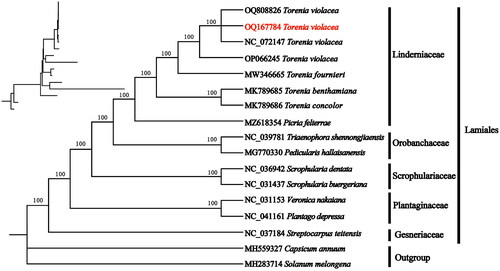
Figure 3. Phylogenetic relationships of Torenia violacea and other species of Lamiales inferred from ML analyses based on the whole chloroplast genome, with Solanum macrocarpon and Capsicum annuum as the outgroup. The numbers on each branches indicate the boot support value of the ML analyses. The sequences used for tree construction are as follows: T. violacea (OQ808826, N_C_072147, OP066245), T. fournieri (MW346665), T. concolor (MK789685, Cheng, Li, et al. Citation2019), T. benthamiana (MK789686, Cheng, Liu, et al. Citation2019), Picria felterrae (MZ618354), Pedicularis hallaisanensis (MG770330, Cho et al. Citation2018), Triaenophora shennongjiaensis (NC_039781, Xia and Wen Citation2018), Scrophularia buergeriana (NC_031437, Yi and Kim Citation2016), Scrophularia dentata (NC_036942), Veronica nakaiana (NC_031437, Choi et al. Citation2016), Plantago depressa (NC_041161), Streptocarpus teitensis (NC_037184, Zhao et al. Citation2019), C. annuum (MH559327, D'Agostino et al. Citation2018), and S. macrocarpon (MH283714, Aubriot et al. Citation2018).
Supplemental Material
Download MS Word (327.2 KB)Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OQ167784. The associated BioProject, SequenceRead Archive, and BioSample numbers are PRJNA918330, SRR22967894, and SAMN32573999.
