Figures & data
Figure 1. Field photos of Campsis radicans. The author Liqiang Wang shot the photo at the position of 35o 15' 9.27" N, 115o 29' 44.44" E. Main identifying traits: 9 ∼ 11 leaflets, calyx five isolobes, shallow in the division, lobed to about one-third. The corolla is slender funnel-shaped, with distinct brownish-red longitudinal stripes inside, and the tube is three times the size of the calyx.

Figure 2. Gene map of the complete chloroplast genome of Campsis radicans. The species name is shown in the top left corner. The map contains six tracks by default. From the center outward, the first track shows dispersed repeats, including direct and palindromic repeats connected by red and green arcs. The second track displays long tandem repeats as blue bars, while the third displays short tandem repeats or microsatellite sequences as differently colored short bars. These colors correspond to the type and description of each repeat, with black representing complex repeats, green for repeat unit size 1, yellow for size 2, purple for size 3, blue for size 4, orange for size 5, and red for size 6. The fourth track displays the SSC, IRa, IRb, and LSC regions. The fifth track shows the GC content along the genome, while the sixth track sounds the genes. The gene names are followed by optional information about codon usage bias and color-coded based on their functional classification. The inner genes are transcribed clockwise, and the outer genes are transcribed anticlockwise. The functional type of the genes is shown in the bottom left corner.
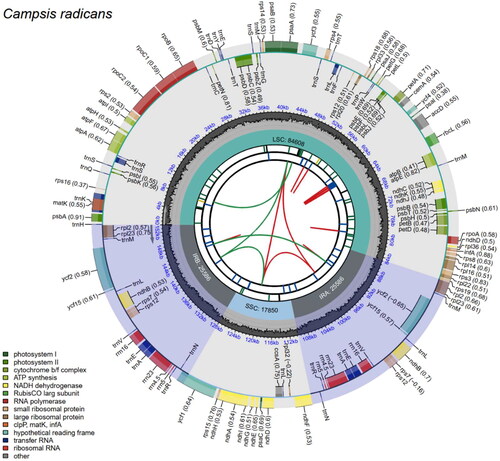
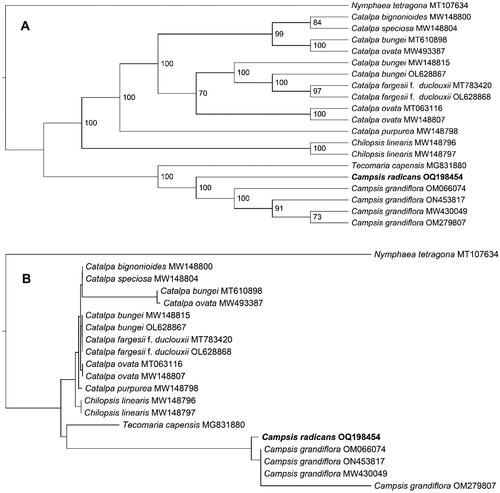
Figure 3. The Maximum-Likelihood phylogeny of Campsis radicans and its close relatives using whole genome sequences. The bootstrap values based on 1000 replicates were shown on each node in the cladogram tree (A). The corresponding phylogram tree is shown in panel B. We downloaded 19 Campsis species plastomes from GenBank, C. grandiflora (Thunb.) K.Schum. (MW430049) (Chen et al. Citation2022), C. grandiflora (OM066074) (Ngai et al. Citation2023), C. grandiflora (OM279807), C. grandiflora (ON453817), Catalpa bignonioides walter (MW148800) (Dong et al. Citation2022), Catalpa bungei C.A.Mey. (MT610898), Catalpa bungei (MW148815) (Dong et al. Citation2022), Catalpa bungei (OL628867) (Li et al. Citation2022), Catalpa fargesii f. duclouxii (dode) Gilmour (MT783420) (Ma et al. Citation2020), Catalpa fargesii f. duclouxii (OL628868) (Li et al. Citation2022), Catalpa ovata G.Don (MT063116) (Wang et al. Citation2020), Catalpa ovata (MW148807) (Dong et al. Citation2022), Catalpa ovata (MW493387), Catalpa purpurea griseb. (MW148798) (Dong et al. Citation2022), Catalpa speciosa teas (MW148804) (Dong et al. Citation2022), chilopsis linearis (cav.) sweet (MW148796) (Dong et al. Citation2022), Catalpa linearis (MW148797) (Dong et al. Citation2022), Tecomaria capensis (Thunb.) spach (MG831880) (Fonseca and Lohmann Citation2018). Nymphaea tetragona Georgi (MT107634) (Sun et al. Citation2021), from the nymphaeaceae, served as the outgroup. The new C. radicans plastome in this study were labeled in bold font.
Supplemental Material
Download MS Word (136.6 KB)Supplemental Material
Download MS Word (751.5 KB)Supplemental Material
Download MS Word (175.9 KB)Data availability statement
The complete chloroplast genome sequence of C. radicans in this study has been submitted to the NCBI database under the accession number OQ198454. https://www.ncbi.nlm.nih.gov. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA928567, SAMN32935701, and SRR23250959.
