Figures & data
Figure 1. The picture of the collected Ocimum basilicum var. basilicum sample. This image shows the whole plant (A), inflorescence (B), and herbarium specimen (C), which were recorded by the authors (Kirankumar S.I. and Raju Balaji).

Figure 2. Circular map of the chloroplast genome of O. basilicum var. basilicum. From the center going outward, the first circle shows the distribution of the repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle displays the tandem repeats marked with short bars. The third circle shows the LSC, SSC, IRa, and IRb regions. The fourth circle shows the percent of GC content. The next circle shows the genes having different colors based on the functional groups. The functional classification is shown at the bottom left. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counter-clockwise direction.
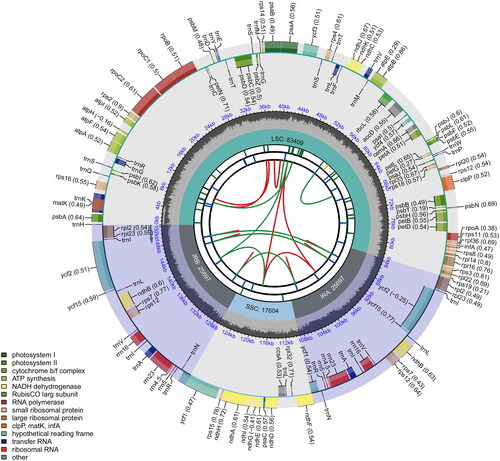
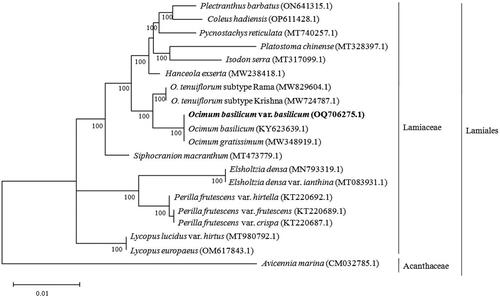
Figure 3. Phylogenetic tree constructed by maximum-likelihood (ML) analysis based on complete chloroplast genome sequences, including O. basilicum var. basilicum (OQ706275.1) sequenced in this study. The numbers on the nodes indicate bootstrap values with 1000 replicates. The sequences used for tree construction are as follows: Ocimum basilicum (KY623639.1; Rabah et al. Citation2017), O. gratissimum (MW348919.1; Balaji et al. Citation2021), O. tenuiflorum subtype Rama (MW829604.1; Harini et al. Citation2021), O. tenuiflorum subtype Krishna (MW724787.1; Kavya et al. Citation2021), Platostoma chinense (MT328397.1), Pycnostachys reticulata (MT740257.1; Wu et al. Citation2021), Plectranthus barbatus (ON641315.1), Coleus hadiensis (OP611428.1), Hanceola exserta (MW238418.1; Zhu et al. Citation2023), Isodon serra (MT317099.1; Zhang et al. Citation2020), Siphocranion macranthum (MT473779.1; Zhao et al. Citation2021), Lycopus lucidus var. hirtus (MT980792.1; Wang et al. Citation2021), Lycopus europaeus (OM617843.1), Elsholtzia densa (MN793319.1; Fu et al. Citation2020), Elsholtzia densa var. ianthina (MT083931.1; Yang et al. Citation2020), Perilla frutescens var. hirtella (KT220692.1), P. frutescens var. frutescens (KT220689.1), P. frutescens var. crispa (KT220687.1), and Avicennia marina (CM032785.1; Natarajan et al. Citation2021).
Supplemental Material
Download TIFF Image (3.7 MB)Supplemental Material
Download TIFF Image (13.3 MB)Supplemental Material
Download TIFF Image (9.1 MB)Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/). The complete chloroplast genome of Ocimum basilicum var. basilicum was deposited in GenBank under the accession OQ706275 (https://www.ncbi.nlm.nih.gov/nuccore/OQ706275). The associated NGS sequencing data files are available from the BioProject, Bio-Sample, and SRA submission under the accession numbers PRJNA949446, SAMN33944295, and SRR23991168, respectively.
