Figures & data
Figure 1. Specimen of S. cinerearia was collected from the Xiangshan Mountain, Huaibei City, China. Photographed by Jun Li on 18 March 2023.

Figure 2. Complete mitochondrial genome map of S. cinerearia with 13 protein coding genes, 22 tRNAs, 2 rRNAs, and an AT-rich region. Outside and inner circles represent the J- and M-strand respectively. Bold lines in circles represent strands in which the genes lie. (Green: tRNAs; yellow: PCGs; greenish yellow: rRNAs; grey: overlaps; white: interval spaces).
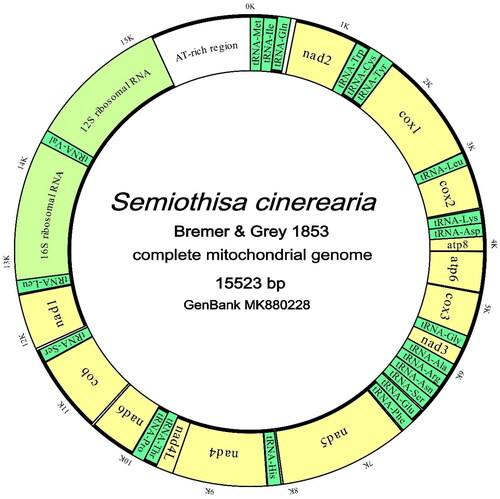
Table 1. Annotations of the mitochondrial genome of S. cinerearia (GenBank MK880228).
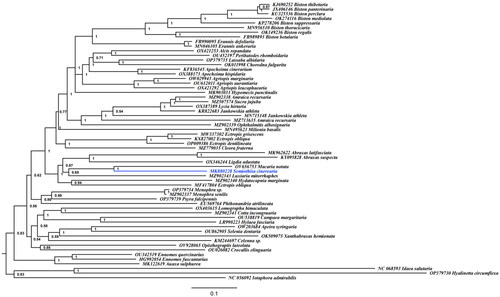
Figure 3. The Bayesian inference (BI) phylogenetic tree was reconstructed based on published mitochondrial genome sequences of subfamily Ennominae and two species of subfamily Geometrinae were used as outgroup. GenBank accession numbers were indicated before the species names. Numbers at the nodes indicated Bayesian posterior probability values.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MK880228. The associated BioProject and Bio-Sample numbers are PRJNA946324 and SAMN33819875, respectively. This mitochondrial genome was not obtained by NGS and does not show the SRA number here.
