Figures & data
Figure 1. Impatiens huangyanensis Jin and Ding Citation2002. (A) Flower lateral view; (B) flower front view; (C) fruits; (D) natural habitat of I. huangyanensis. All the photos were taken by Ming Jiang.

Figure 2. The chloroplast genome of Impatiens huangyanensis. The map contains six tracks. From the center outward, the first track shows the dispersed repeats, which consist of direct and palindromic repeats, connected with red and green arcs. The second track indicates the long tandem repeats as short blue bars. The third track reveals the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, type of repeat they represent, and the description of the repeat types are as follows: black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). the chloroplast genome contains an LSC region, an SSC region, and two IR regions, and they are shown on the fourth track. The GC content along the genome is shown on the fifth track. Genes are color-coded according to their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The bottom left corner indicates the key for the functional classification of the genes.
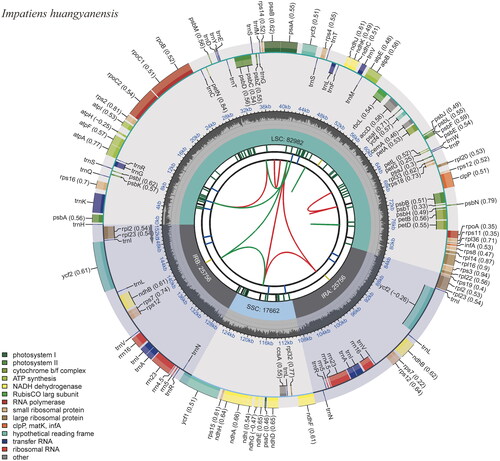
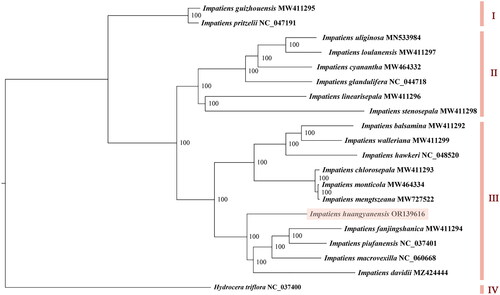
Figure 3. The maximum-likelihood tree based on complete chloroplast genome sequences of Impatiens huangyanensis and 18 other Impatiens species, with Hydrocera triflora as the outgroup species. The numbers next to the nodes show bootstrap support values. The phylogenetic tree was generated by PhyML 3.1 with the maximum-likelihood method.
Supplemental Material
Download MS Word (567.5 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ OR139616. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA897866, SRR22178406, and SAMN31582508, respectively.
