Figures & data
Figure 1. Neolissochilus soroides was collected from Daying River, Tengchong city, Yunnan province, China (24°36′36ʺ N, 97°49′12ʺ E) (photo by Ye Chen).

Figure 2. Mitochondrial genome map of Neolissochilus soroides. H-strand is located in the outer ring and L-strand is located in the inner ring. The gene consists of 13 protein-coding genes, two rRNA genes, 22 tRNA genes and three non-coding control regions. Among them, yellow: complex I (NADH dehydrogenase); light green: complex III (ubiquinol cytochrome c reductase); pink: complex IV (cytochrome c oxidase); dark green: ATP synthase; blue: transfer RNAs; red: ribosomal RNAs.
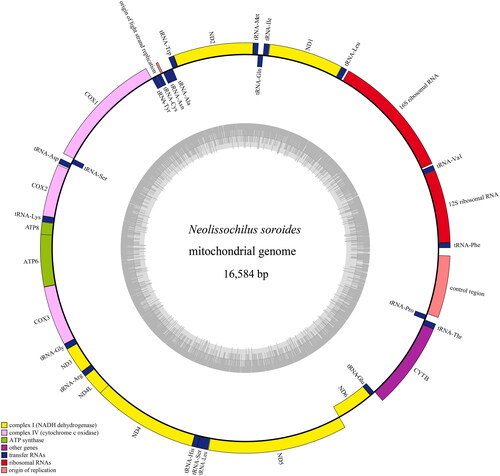
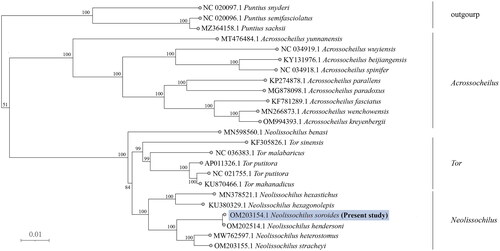
Figure 3. Phylogenetic analysis of N. soroides based on the entire mtDNA genome sequences of 24 Cypriniformes available in GenBank. Numbers above the nodes indicate 1000 bootstrap values. Accession numbers are shown before species names. The following sequences were used: Neolissochilus hendersoni OM202514.1 (Guo et al. Citation2023), Neolissochilus stracheyi OM203155.1 (Wei, Z. et al. 2022), Neolissochilus heterostomus MW762597.1 (He et al. Citation2021), Neolissochilus hexagonolepis KU380329.1 (Zhou et al. Citation2016), Neolissochilus hexastichus voucher MN378521.1 (Singh et al. Citation2021), Tor sinensis KF305826.1 (Huang et al. Citation2015), Tor malabaricus NC 036383.1 (Chandhini et al. Citation2019), Tor putitora AP011326.1 (Sati et al. Citation2014), Tor mosal mahanadicus KU870466.1 (Sarma et al. Citation2022), Tor putitora NC 021755.1 (Sati, J. et al. 2014), Neolissochilus benasi MN598560.1 (Gu et al. Citation2020), Acrossocheilus yunnanensis MT476484.1 (Chen et al. Citation2022), Acrossocheilus spinifer NC 034918.1 (Zhao et al. Citation2022), Acrossocheilus beijiangensis KY131976.1 (Liu et al. Citation2018), Acrossocheilus wuyiensis NC 034919.1 (Yuan et al. Citation2017), Acrossocheilus paradoxus MG878098.1 (Ju et al. Citation2018), Acrossocheilus parallens KP274878.1 (Xie et al. Citation2016), Acrossocheilus fasciatus KF781289.1 (Cheng et al. Citation2015), Acrossocheilus kreyenbergii OM994393.1 (Zhou et al. Citation2023), Acrossocheilus wenchowensis MN266873.1 (Pan et al. Citation2019), Puntius snyderi NC 020097.1 (Jang-Liaw et al. Citation2013), Puntius sachsii MZ364158.1 (Sun et al. Citation2023), Puntius semifasciolatus NC 020096.1 (Sun et al. Citation2023), Puntius chalakkudiensis NC 018566.1 (Khare et al. Citation2014).
Supplemental Material
Download MS Word (79.1 KB)Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/OM203154.1/) under accession no. OM203154. The associated BioProject, SRA, and Bio-Sample Numbers are PRJNA817098, SRR18356122, and SAMN26747790.
