Figures & data
Figure 1. The morphology of leaf of S. sagittifolia. (a) The basal leaves of S. sagittifolia. (b) The leaves abaxially of S. sagittifolia. Leaf blades in middle and upper stem triangular, lanceolate, linear to subulate, apex long-caudate, base attenuate, margin with a few small sharp teeth to subentire. Leaves abaxially usually dark purple, capitula fewer, and involucres narrowly campanulate. The photos of S. sagittifolia were taken by the Zhi-Xi Fu in Guangwushan Mountain, Nanjiang county, Bazhong city, Sichuan province, China (the voucher number: Ya Deng, DY158).

Figure 2. The chloroplast genome map of S. sagittifolia generated using CPGview. Boxes of different sizes and colors in the outermost circle represent genes and their lengths. Genes inside the circle are transcribed clockwise, and those on the outside are transcribed counter-clockwise. The grey area in the middle circle represents the variation of GC content at different positions, and the regions and lengths represented by the tetrameric structures (LSC, SSC, IRa, and IRb) are plotted in different colors on the inner circle.
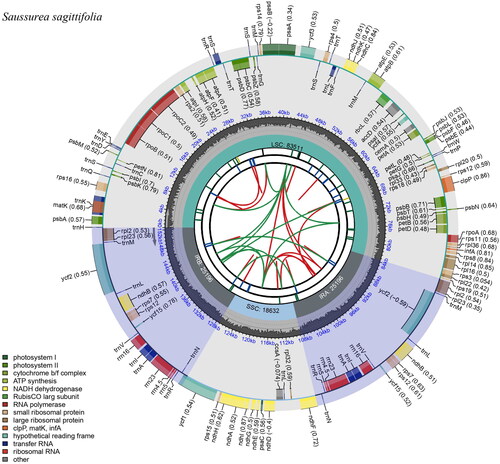
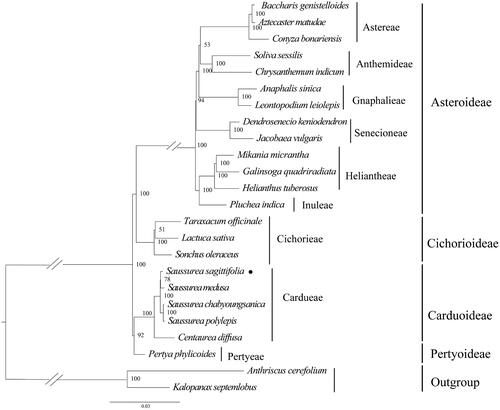
Figure 3. Maximum-likelihood phylogeny of S. sagittifolia and related taxa based on 24 complete chloroplast genomes. Anthriscus cerefolium (Apiaceae), Kalopanax septemlobus (Araliaceae) were used as outgroups. The maximum-likelihood bootstrap support values are along the branches. Circles represent newly sequenced species. The following sequences were used: Aztecaster matudae KX063935 (Vargas et al. Citation2017), Baccharis genistelloides KX063864 (Vargas et al. Citation2017), Conyza bonariensis KX792499 (Wang et al. Citation2018), Chrysanthemum indicum JN867589 (unpublished), Soliva sessilis KX063863 (Vargas et al. Citation2017), Anaphalis sinica KX148081 (unpublished), Leontopodium leiolepis KM267636 (unpublished), Dendrosenecio keniodendron KY434193 (unpublished), Jacobaea vulgaris HQ234669 (Doorduin et al. Citation2011), Galinsoga quadriradiata KX752097 (Wang et al. Citation2018), Mikania micrantha KX154571 (Huang et al. Citation2016), Helianthus tuberosus MG696658 (unpublished), Pluchea indica MG452144 (Zhang et al. Citation2017), Taraxacum officinale KU361241 (unpublished), Lactuca sativa AP007232 (unpublished), Sonchus oleraceus MG878405 (Hereward et al. Citation2018), Saussurea sagittifolia ON094066 (in this study), Saussurea polylepis MF695711 (Seon et al. Citation2017), Centaurea diffusa KJ690264 (unpublished), Pertya phylicoides MN935435 (Wang et al. Citation2020), Anthriscus cerefolium GU456628 (Downie and Jansen Citation2015), and Kalopanax septemlobus NC022814 (Li et al. Citation2013).
Supplemental Material
Download MS Word (309.5 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON094066. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA939009, SRR23682417, and SAMN33558826, respectively.
