Figures & data
Figure 1. Reference images of Acalypha hispida. A. The plant morphology of A. hispida, a much-branched shrub with female inflorescences that are spicate and axillary; B. The morphology of inflorescences and leaves in A. hispida, includes leaf blades up to 20 × 15 cm, with serrations on the margins, solitary female inflorescences that are dense-flowered and bright red, and an inflorescence rachis that is sparsely pubescent. These pictures were taken by a local professional team in xishuangbanna tropical botanical garden (XTBG) of chinese academy of sciences (CAS), located at 101° 25' E, 21° 41' N, altitude: 570 m, Yunnan Province, China.

Figure 2. The chloroplast genome map of A. hispida. Genes shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. Genes belonging to different functional groups are displayed in different colors. The darker gray area in the inner circle indicates the GC content, and the lighter gray indicates the content of the chloroplast genome.
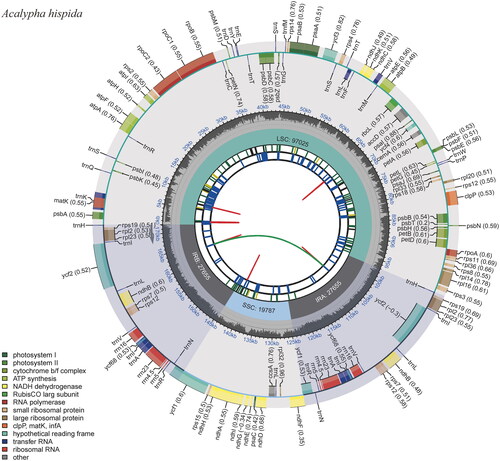
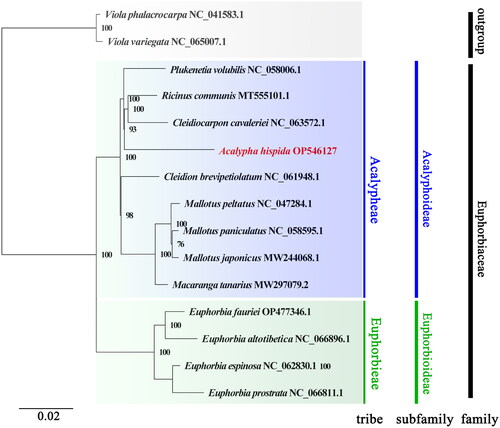
Figure 3. Phylogenetic analysis based on the complete chloroplast genomes. The bootstrap values are shown at the nodes, and the species and GenBank accession numbers are shown at the end of each branch. The phylogenetic tree of A. hispida inferred with the maximum-likelihood (ML) method based on the concatenated complete chloroplast genome sequence, with Viola phalacrocarpa and Viola variegate as outgroups. GenBank accession numbers of the following sequences were used: Cleidiocarpon cavaleriei, NC_063572.1 (Xin et al. Citation2019); Cleidion brevipetiolatum, NC_061948.1; Euphorbia altotibetica, NC_066896.1; Euphorbia espinosa, NC_062830.1; Euphorbia fauriei, OP477346.1; Euphorbia prostrata, NC_066811; Macaranga tanarius, MW297079.2 (Tang et al. Citation2021); Mallotus japonicus, MW244068.1 (Ke et al. Citation2020); Mallotus paniculatus, NC_058595.1 (Ke et al. Citation2020); Mallotus peltatus, NC_047284.1; Plukenetia volubilis, NC_058006.1 (Hu et al. Citation2018); Ricinus communis, MT555101.1 (Hu et al. Citation2018); Viola phalacrocarpa, NC_041583.1 (Cai et al. Citation2020); and Viola variegata, NC_065007.1.
Supplemental Material
Download PDF (303.3 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank of the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov. The complete chloroplast genome has been deposited in GenBank under the accession no. OP546127. And the associated Bio-project, SRA, Bio-sample numbers are PRJNA884882, SRX17729733 and SAMN31058094 respectively.
