Figures & data
Figure 1. Photos of S. marina from a coastal region in Buan-gun, Jeollabuk-do, South Korea. (A) Whole plant at the young growing stage. Stems erect to ascending or prostrate, usually much-branched proximally. (B) Flower with pink petals at the mature stage. Sepal has glandular hairy. Photos were taken by Khongorzul Erdenebaatar.

Figure 2. Physical map of the S. marina chloroplast genome. The map was generated by CPGView. Genes located on the inner and outer of circle are transcribed clockwise and anticlockwise, respectively. The dark grey inner circle indicates GC content. Large single-copy (LSC), small single-copy (SSC), and inverted repeats (IRA and IRB) are indicated in the inner layer. The functional classification of the genes is provided in the bottom left corner.
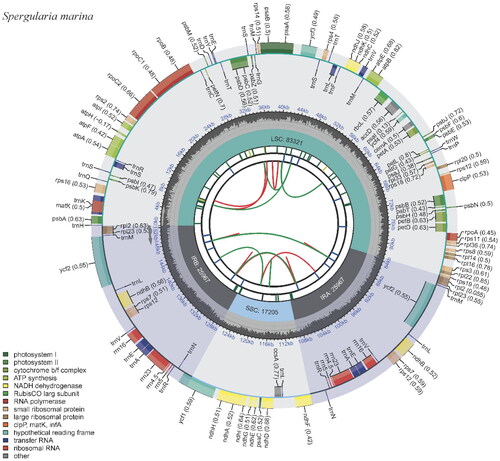
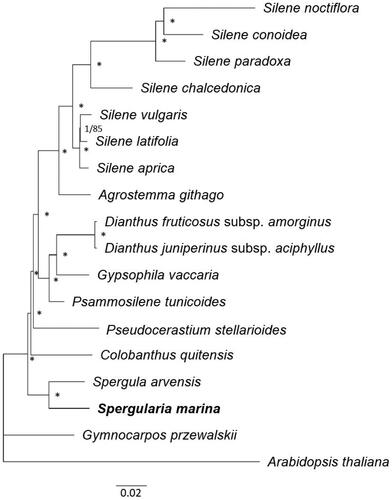
Figure 3. Phylogenetic relationships among 17 Caryophyllaceae species based on complete chloroplast genome sequences. Arabidopsis thaliana was used as an outgroup. Phylogenetic trees were constructed using ML and BI methods. Note that both methods yielded the same tree topology. Numbers on node indicate BI posterior probabilities and ML bootstrap values (%), respectively. The posterior probabilities (1.0) and bootstrap support (100%) are represented with an asterisk. Sequences used for tree construction were as follows: Agrostemma githago (KF527884, Sloan et al. Citation2014), Colobanthus quitensis (NC_028080), Dianthus fruticosus subsp. amorginus (MT150102), Dianthus juniperinus subsp. aciphyllus (MT150107), Gymnocarpos przewalskii (MF795140), Gypsophila vaccaria (NC_040936, Yao et al. Citation2019), Psammosilene tunicoides (NC_045947. Li et al. Citation2019), Pseudocerastium stellarioides (MT507771, Yao et al. Citation2021), Silene aprica (NC_040934, Yao et al. Citation2019), Silene chalcedonica (NC_023359, Sloan et al. Citation2014), Silene conoidea (NC_023358, Sloan et al. Citation2014), Silene latifolia (NC_016730, Sloan et al. Citation2012), Silene noctiflora (NC_016728, Sloan et al. Citation2012), Silene paradoxa (NC_023360, Sloan et al. Citation2014), Silene vulgaris (NC_016727, Sloan et al. Citation2012), Spergula arvensis (NC_041240, Yao et al. Citation2019), and Spergularia marina (OR038716). Arabidopsis thaliana (MK380721, Park et al. Citation2020) was used as an outgroup. The scale bar represents the number of substitutions per site.
Supplemental Material
Download MS Word (278.1 KB)Data availability statement
The chloroplast genome sequence data that support the findings of this study are freely available in GenBank of NCBI https://www.ncbi.nlm.nih.gov/ under the accession number OR038716. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835661, SRR25000439, and SAMN35847521, respectively.
