Figures & data
Figure 1. The morphology of T. marina (A) in flasks and (B) under a microscope. Photograph was taken by Fangfang Yang. It is a unicellular with a size range between 10 and 20 μm.
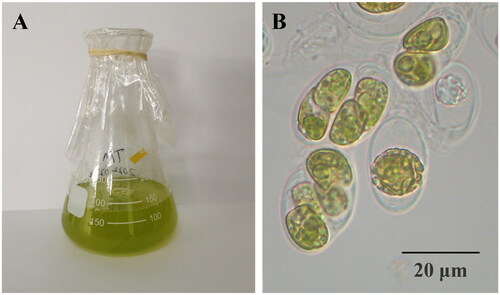
Figure 2. Gene map of the complete chloroplast genome of T. marina using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). Genes shown on the outside of the circle are transcribed clockwise, while those inside are transcribed counterclockwise. Arrangement of 125 genes represented in the map, including 81 protein-coding genes, six rRNA, and 38 tRNA genes.
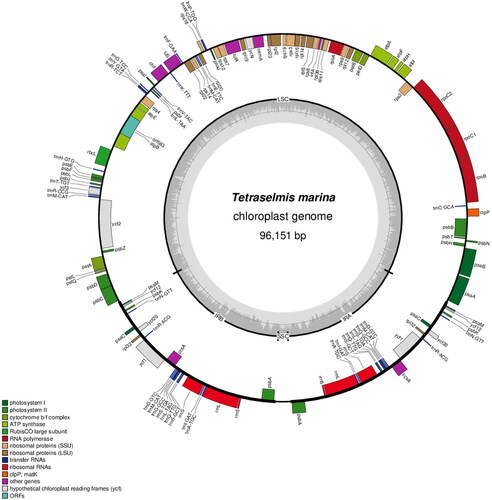
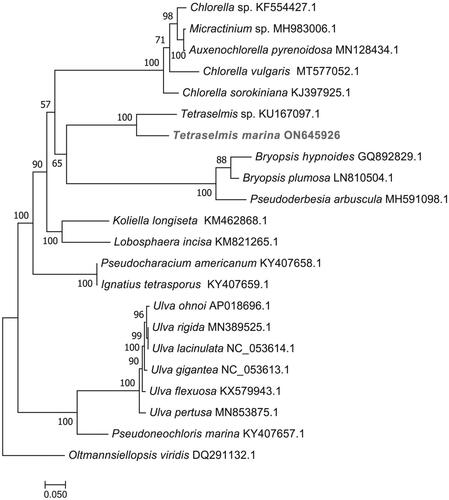
Figure 3. Maximum-likelihood (ML) phylogenetic tree based on complete chloroplast genomes. Oltmannsiellopsis viridis was used as an outgroup species. The numbers on each branches indicate the boot support value of the ML analyses. The scale bar was 0.050. The following sequences were used: Chlorella sp. KF554427.1, Micractinium sp. MH983006.1, Auxenochlorella pyrenoidosa MN128434.1, Chlorella vulgaris MT577052.1, Chlorella sorokiniana KJ397925.1, Tetraselmis sp. CCMP 881 KU167097.1 (Turmel et al. Citation2020), Bryopsis hypnoides GQ892829.1, Bryopsis plumosa GenBank: LN810504.1, Pseudoderbesia arbuscula MH591098.1, Koliella longiseta KM462868.1, Lobosphaera incisa KM821265.1, Pseudocharacium americanum KY407658.1, Ignatius tetrasporus KY407659.1, Ulva ohnoi AP018696.1, Ulva rigida MN389525.1, Ulva lacinulata NC_053614.1, Ulva gigantea NC_053613.1, Ulva flexuosa KX579943.1, Ulva pertusa MN853875.1, Pseudoneochloris marina KY407657.1, and Oltmannsiellopsis viridis DQ291132.1 (Pombert et al. Citation2006).
Supplemental Material
Download MS Word (1.3 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON645926. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA844566, SRR19521163, and SAMN28834016, respectively.
