Figures & data
Figure 1. Specimen of Zygophyllum brachypterum (this unpublished photo, taken in Aksu Prefecture, Xinjiang Uygur Autonomous Region of China by Prof. Peipei Jiao, is used with permission), Stems of Z. brachypterum are much branched and tender, and leaves have two leaflets that are oblong to oblanceolate, thin, and apex obtuse. Petioles are equal to or shorter than leaflets.

Figure 2. The complete chloroplast genome map of Z. brachypterum. The quadripartite structure is marked with LSC, SSC, and IRA/IRB. Different functional genes are represented by bars with different colors. The gray part in the inner circle indicates the GC content. Genes outside the circle are transcribed in the clockwise direction, and those inside the circle are transcribed in the counterclockwise direction.
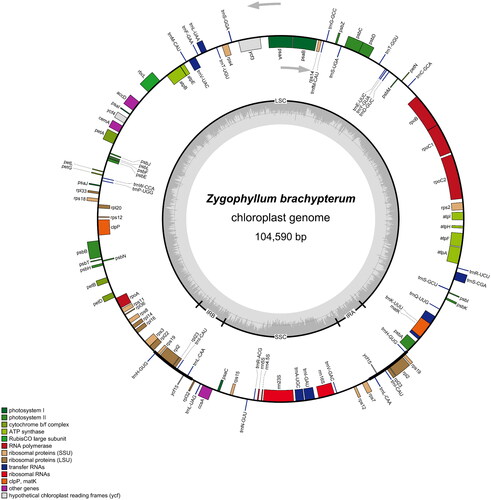
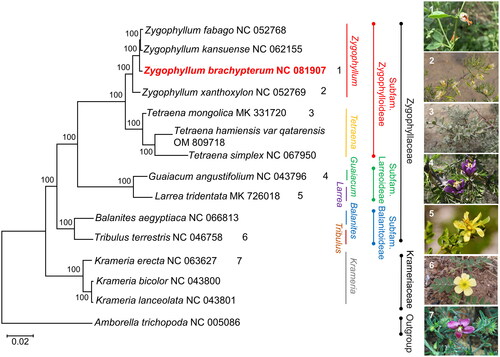
Figure 3. The phylogenetic tree is based on the common PCGs of 14 related species from Krameriaceae and Zygophyllaceae. A. trichopoda is regarded as the outgroup. The bootstrap values based on 1000 replications are exhibited at the branches. Scale bar = 0.02. The images of Z. brachypterum, Z. xanthoxylum, and T. mongolica were taken from Polat Muhtar and Ningmei Chen with permission for use. The images of Guaiacum angustifolium, Larrea tridentata, Tribulus terrestris, and Krameria erecta were downloaded from the public domain and can be used without asking permission as declared by the copyright holders. The following sequences downloaded from NCBI were used: Zygophyllum fabago NC_052768 (Xu et al. Citation2020), Zygophyllum kansuense NC_062155 (Wang et al. Citation2022a), Zygophyllum xanthoxylum NC_052769 (Xu et al. Citation2020), Tetraena mongolica MK331720 (Wang et al. Citation2022b), Tetraena hamiensis var. qatarensis OM809718 (Ahmad et al. Citation2023), Tetraena simplex NC_067950 (Ahmad et al. Citation2023), Guaiacum angustifolium NC_043796 (Gonçalves et al. Citation2019b), Larrea tridentata MK726018 (Gonçalves et al. Citation2019a), Balanites aegyptiaca NC_066813 (Al-Juhani et al. Citation2022), Tribulus terrestris NC_046758 (Yan et al. Citation2019), Krameria erecta NC_063627 (Banerjee et al. Citation2022), Krameria bicolor NC_043800 (Gonçalves et al. Citation2019b), Krameria lanceolata NC_043801 (Gonçalves et al. Citation2019b), and Amborella trichopoda NC_005086 (Goremykin et al. Citation2003).
Supplemental Material
Download TIFF Image (1.1 MB)Supplemental Material
Download TIFF Image (6.6 MB)Supplemental Material
Download TIFF Image (2.2 MB)Supplemental Material
Download TIFF Image (2.6 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number NC_081907. The associated BioProject, SRA, and BioSample numbers are PRJNA952865, SRR24097901, and SAMN34090572, respectively.
