Figures & data
Figure 1. Species image of T. nanpanjiangensis. The most characteristic features of this species are dorsal fin III-7; anal fin III-5; pectoral fin I-9; ventral fin I-6; caudal fin 16; normal eyes; and caudal fin slightly forked. Photo taken by Zhiqiang Liang in changsha, China.

Figure 2. Map of the assembled T. nanpanjiangensis mitochondrial genome (GenBank Accession: OQ274895) consisting of 13 protein-coding genes (dark green, light green and yellow), 22 transferRNAs genes (dark blue), two ribosomal RNA genes (red), and one non-coding control region (D-loop, light blue). genes encoded on the reverse strand and forward strand are illustrated outside the circle and inside the circle, respectively. The inner ring displays the GC content of the genome.
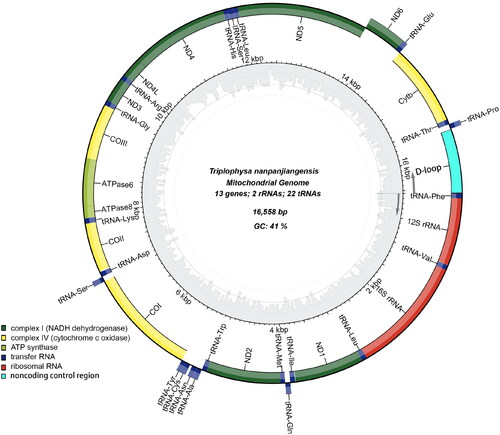
Table 1. Characteristics of the mitochondrial genome of T. nanpanjiangensis.
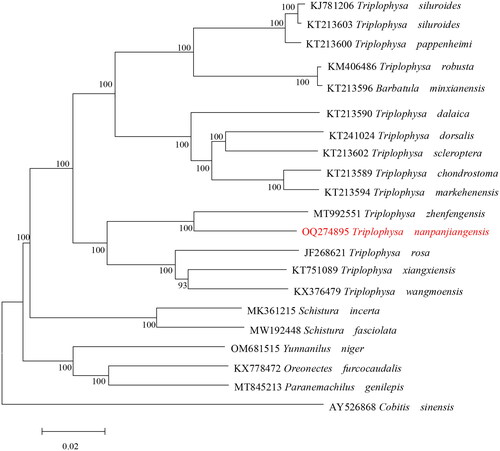
Figure 3. Maximum-likelihood (ML) phylogenetic tree was reconstructed based on the complete mitochondrial genome of T. nanpanjiangensis and other 20 Cypriniformes fishes. Accession numbers were indicated after the species names. Numbers at the nodes indicated bootstrap support values from 1000 replicates under the GTR + F + I + G4 model.
Supplemental Material
Download MS Word (166.6 KB)Data availability statement
The consensus genome sequence is openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. OQ274895. The associated BioProject, SRA and BioSample numbers are PRJNA939422, SRR23646241 and SAMN33476296, respectively.
