Figures & data
Figure 1. Species image of Z. mairei. (A) Morphology of the whole plant. Z. mairei is a spinose tree of 15 m in height. (B) Morphological characteristics of leaves of Z. mairei. Leaves papery, ovate-lanceolate, and apically long acuminate. The photograph was taken by the corresponding author Yuan Huang at Kunming Botanic Garden, Yunnan Province of China (25°14'56"N, 102°74'07"E), on March 2023.

Figure 2. Genomic map of overall features of the chloroplast genome of Z. mairei by CPGview. The map contains six tracks from the center outward. The first track represents the dispersed repeats, the red arcs represent the direct repeats and the green arcs represent the palindromic repeats. The second track is a long tandem repeat marked by short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars of different colors. The fourth track is the tetrameric structure of the chloroplast genome, containing LSC, IRa, SSC, and IRb. The fifth track is the GC content of the chloroplast genome. The last track is coding genes categorized according to function. The transcription directions for the inner and outer genes are clockwise and anti-clockwise, respectively.
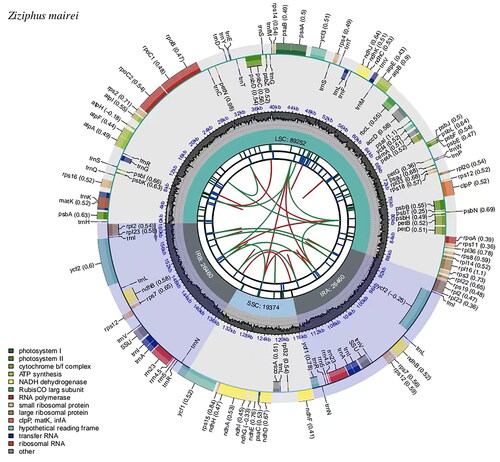
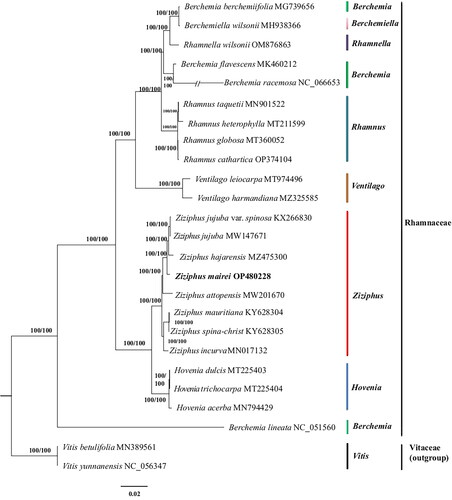
Figure 3. A phylogenetic tree was constructed using the maximum-likelihood method based on complete chloroplast sequences from 23 Rhamnaceae species and 2 Vitaceae species as outgroups. The following sequences were used: Berchemia berchemiifolia MG739656 (Kyeong et al. Citation2018), berchemiella wilsonii MH938366(Li et al. Citation2019), Rhamnella wilsonii OM876863, Berchemia flavescens MK460212 (Zhu et al. Citation2019), Berchemia racemosa NC_066653 (Park and Koo Citation2023), Rhamnus taquetii MN901522 (Jin et al. Citation2020), Rhamnus heterophylla MT211599 (Li et al. Citation2020), Rhamnus globosa MT360052 (Xie et al. Citation2020), Rhamnus cathartica OP374104 (Shi et al. Citation2023), Ventilago leiocarpa MT974496 (Lu et al. Citation2021), Ventilago harmandiana MZ325585 (Wanichthanarak et al. Citation2023), Z. jujuba var. spinosa KX266830 (Kyeong et al. Citation2018), Z. jujuba MW147671 (Huang et al. Citation2017), Z. hajarensis MZ475300 (Asaf et al. Citation2022), Ziziphus attopensis MW201670 (Li Citation2021), Ziziphus mauritiana KY628304 (Huang et al. Citation2017), Ziziphus spina-christ KY628305 (Huang et al. Citation2017), Ziziphus incurva MN017132 (Wang et al. Citation2019), Hovenia dulcis MT225403 (Liu et al. Citation2021), Hovenia trichocarpa MT225404 (Li et al. Citation2020), Hovenia acerba MN794429 (Zhang et al. Citation2020), Berchemia lineata NC_051560 (Xie et al. Citation2020), Vitis betulifolia MN389561 (Xu and Xu Citation2021), Vitis yunnanensis NC_056347 (Xu and Xu Citation2021).
Supplemental Material
Download MS Word (444.8 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under accession no. OP480228.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA882268, SRR21639213, and SAMN30930803, respectively.
