Figures & data
Figure 1. Reference image of Huperzia arctica. Perennial plants, 5–10 cm tall, sporangia are yellow and produced at the leaf base, and spores are yellow as well. Bulbils are located at the apices of stems. The photograph was taken by Youngsim Hwang in Ny-Ålesund, Svalbard (78°54′49.0″ N; 11°58′53.6″ E) on 17 July 2022.

Figure 2. Map of the chloroplast genome of Huperzia arctica. Genes lying outside the outer circle are transcribed clockwise, while those inside the circle are transcribed counterclockwise. Genes belonging to different functional groups are color-coded. The innermost darker gray corresponds to GC content, while the lighter gray corresponds to at content. IR, inverted repeat; LSC, large single copy region; SSC: small single copy region.
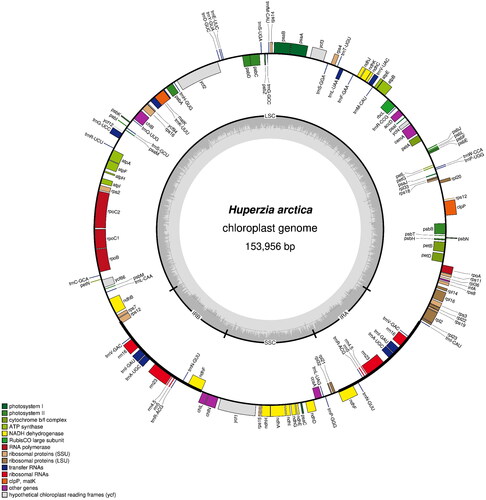
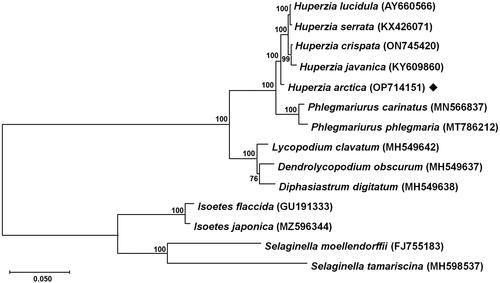
Figure 3. Maximum-likelihood phylogenetic tree based on 14 complete chloroplast genomes. The tree was constructed based on the GTR + G + I model using MEGA11 with nucleotide sequences of 36 conserved orthologous protein-coding genes. The numbers on each internal node indicate bootstrap values based on 1000 iterations. The following sequences were used: Huperzia lucidula (AY660566) (Wolf et al. Citation2005), H. serrata (KX426071) (Guo et al. Citation2016), H. crispata (ON745420) (Yin et al. Citation2022), H. javanica (KY609860) (Zhang et al. Citation2017), H. arctica (OP714151) (this study), Phlegmariurus carinatus (MN566837) (Luo et al. Citation2019), P. phlegmaria (MT786212) (Tang et al. Citation2020), Lycopodium clavatum (MH549642) (Mower et al. Citation2019), Dendrolycopodium obscurum (MH549637) (Mower et al. Citation2019), Diphasiastrum digitatum (MH549638) (Mower et al. Citation2019), Isoetes flaccida (GU191333) (Karol et al. Citation2010), I. japonica (MZ596344) (Lin et al. Citation2022), Selaginella moellendorffii (FJ755183) (Smith Citation2009), S. tamariscina (MH598537) (Zhang et al. Citation2019). Isoetes flaccida, I. japonica, Selaginella moellendorffii, S. tamariscina were used as outgroup taxa. Scale bar refers to a phylogenetic distance of 0.05 nucleotide substitutions per site.
Supplemental Material
Download PDF (148 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP714151. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA941097, SRP425714, and SAMN33589810, respectively.
