Figures & data
Figure 1. Morphological observation of the Metarhizium pinghaense 15 R strain. (A), the strain was cultured on PDA medium at 25 °C for 2 weeks; (B), green peach aphid (Myzus persicae) infected with the strain. The cadaver was covered with the strain’s white hyphae and green conidia and was photographed by InJi Heo at the insect pathology and bioactives laboratory of jeonbuk National university, Korea, on 19 jan 2023.
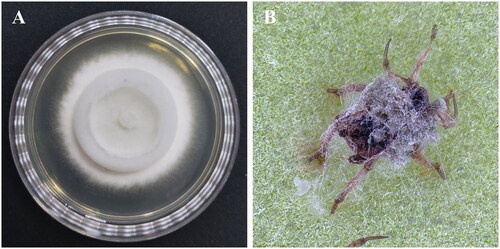
Figure 2. Graphic representation of19 features identified the Metarhizium pinghaense 15 R mitochondrial genome. The map was prepared using OGDRAW program (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). Genes are shown outside, and GC and at contents across the mitochondrial genome are shown with dark and light shading, respectively, inside the inner circle. Genes are color-coded by their functional classification.
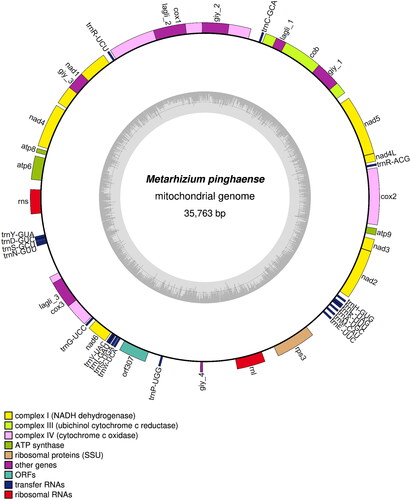
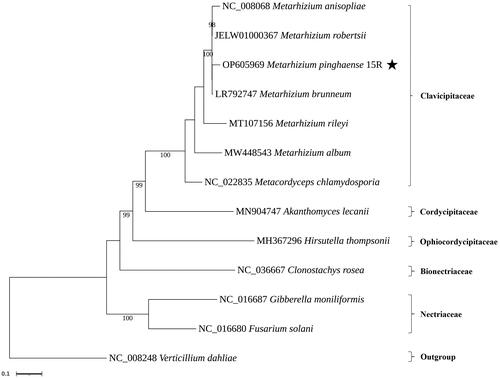
Figure 3. Phylogenetic trees based on the mitochondrial nucleotide sequence in related fungal species. Phylogenetic trees based on the mitochondrial nucleotide sequence in related fungal species. Phylogenetic tree of mitochondrial genomes of Metarhizium pinghaense 15 R and its related 12 species. We used seven species of Clavicipitaceae and representative species of other families with available mitogenomes in Hypocreales. Protein-coding sequences conserved in the mitochondrial genomes of 13 species were multiple-aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate a phylogenetic tree using the maximum likelihood approach as implemented in RAxML v8.2.12 (StamatakisCitation2014). the bootstrap values were indicated for nodes that support values of >70%. and Verticillium dahliae NC_008248.1 (Pantou et al. Citation2006) used an outgroup of this tree. The species under study is highlighted by the star. The following sequences were used: Metarhizium album MW448543.1 (Sun et al. Citation2021), Metarhizium anisopliae NC_008068.1 (Ghikas et al. Citation2006), Metarhizium brunneum LR792747.1 (Kortsinoglou et al. Citation2020), Metarhizium rileyi MT107156.1 (Zhang et al. Citation2020a), Metarhizium robertsii JELW01000367, Akanthomyces lecanii MN904747.1 (Zhang et al. Citation2020b), Clonostachys rosea NC_036667.1, Fusarium solani NC_016680.1 (Al-Reedy et al. Citation2012), Gibberella moniliformis NC_016687.1 (Al-Reedy et al. Citation2012), Hirsutella thompsonii MH367296.1 (Wang et al. Citation2018), Metacordyceps chlamydosporia NC_022835.1 (Lin et al. Citation2015), and Verticillium dahliae NC_008248.1 (Pantou et al. Citation2006).
Supplemental Material
Download MS Word (94 KB)Supplemental Material
Download MS Word (68.3 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OP605969. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA815910, SRR18320042, and SAMN26650411, respectively.
