Figures & data
Figure 1. Image of L. adscendens. Jieqiong Shen collected the plant from Mt. Danan of Hui Lai town, Jieyang city, Guangdong province of China (GPS: E116°10′44.8″, N23°16′44.6″). The floating stem of L. adscendens can be up to 3 meters long and the standing stem up to 60 cm long. A rootlike float with many palpate roots, often clustered in a cylindrical. The leaves are obovate, elliptic or obovate-lanceolate. Branches in xerophilic environments are often puberulent but rarely flower.

Figure 2. Schematic map of overall features of the chloroplast genome of L. adscendens. The genes coding forward are on the outside of the circle, and the genes coding backward are on the inside of the circle. The gray circle inside represents the GC content. Different colors represent different gene types, the detailed gene types are listed in the captions.
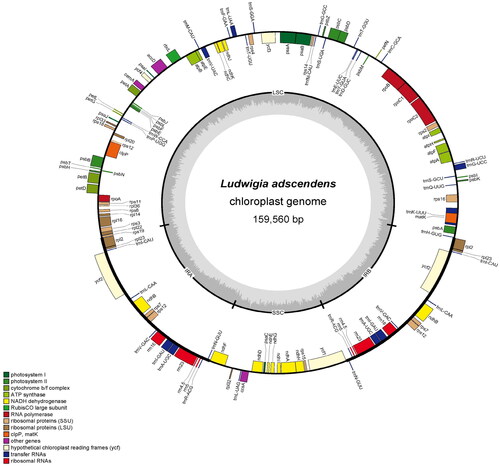
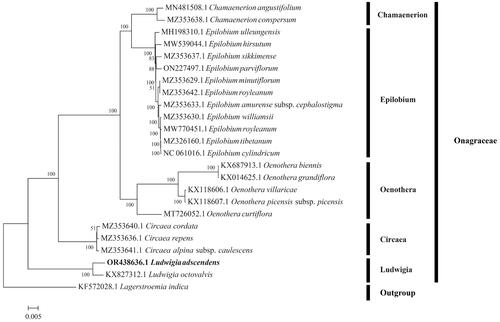
Figure 3. Maximum likelihood tree based on 84 protein-coding gene sequences of 24 complete chloroplast genomes. The GenBank accession number has been provided within parentheses after the species name. Numbers above the branches indicate bootstrap-supporting values based on 1000 replicates. The bar represents 0.005 nucleotide substitution per site.
Supplemental Material
Download MS Word (145.4 KB)Data availability statement
The sequence information of the complete chloroplast genome assembly of the L. adscendens is available on the NCBI website (https://www.ncbi.nlm.nih.gov), with the accession numbers OR438636.1. The associated BioProject, Bio-Sample, and SRA are PRJNA1005061, SAMN36972193, and SRR25627446, respectively.
