Figures & data
Figure 1. Field photo of Acalypha australis. The photo was shot by Liqiang Wang at the position of 35°16′30.0″N, 115°28′41.08″E. Main identifying traits: slender branchlets; leaves membranous, long ovate, subrhomboid ovate or broadly lanceolate; unisexual flowers, inflorescences axillary, thinly terminal; seeds subovate, seed coat smooth. Panel A: Panorama, B: Side profile, C: Bird's-eye view.
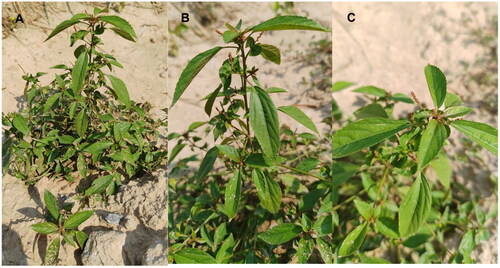
Figure 2. Gene map of the complete chloroplast genome of Acalypha australis. The map contains five tracks by default. From the Center outward, the first track shows dispersed repeats, including direct and palindromic repeats connected by red and green arcs. The second track displays long tandem repeats as blue bars, while the third track shows short tandem repeats or microsatellite sequences as differently colored short bars. These colors correspond to the type and description of each repeat, with black representing complex repeats, green for repeat unit size 1, yellow for size 2, purple for size 3, blue for size 4, orange for size 5, and red for size 6. The fourth track displays the SSC, IRa, IRb, and LSC regions. The fifth track shows the genes. The gene names are followed by optional information about codon usage bias and are color-coded based on their functional classification. The inner genes are transcribed clockwise, and the outer genes are transcribed anticlockwise. The functional classification of the genes is shown in the bottom left corner.
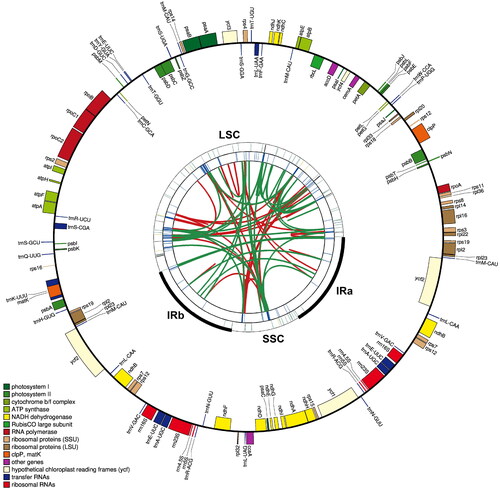
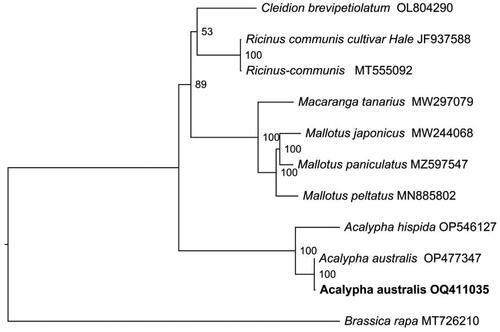
Figure 3. The maximum likelihood phylogeny of Acalypha australis and its close relatives using whole genome sequences. The bootstrap values based on 1000 replicates were shown on each node in the cladogram tree. Ten species are from Euphorbiaceae, which are Mallotus peltatus (MN885802) (Ke et al. Citation2020), Mallotus japonicus (MW244068) (Wu et al. Citation2021), Ricinus communis (MT555092), A. hispida (OP546127), Mallotus paniculatus (MZ597547) (Li et al. Citation2022), Ricinus communis (JF937588) (Rivarola et al. Citation2011), macaranga tanarius (MW297079), cleidion brevipetiolatum (OL804290), A. australis (OP477347) and A. australis (OQ411035, in this study). Brassica rapa (MT726210) (Wu et al. Citation2021), from the brassicaceae, served as the outgroup. The new A. australis plastomes (OQ411035) in this study were labeled in bold font.
Supplemental Material
Download PDF (785.2 KB)Data availability statement
The complete plastome sequence of A. australis in this study has been submitted to the NCBI database (https://www.ncbi.nlm.nih.gov) under the accession number OQ411035. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA928567, SAMN36852646, SRR23328024.
