Figures & data
Figure 1. Species reference image of H. marinum ssp. marinum (voucher no. BTBU20220515) under cultivation are from our laboratory and photography by Suping Yu. (A) Hydroponic culture of H. marinum. (B) Seedlings of H. marinum during the stages of two leaves and one heart.

Figure 2. Circular genetic map of the chloroplast genome of H. marinum ssp. marinum. The map consists of three circles: the inner circle indicates the positions of the LSC, SSC, IRA, and IRB regions, respectively. The Middle circle represents the GC content. The outer circle displays genes of various functional categories, each depicted in different colors as illustrated in the lower left corner of the picture.
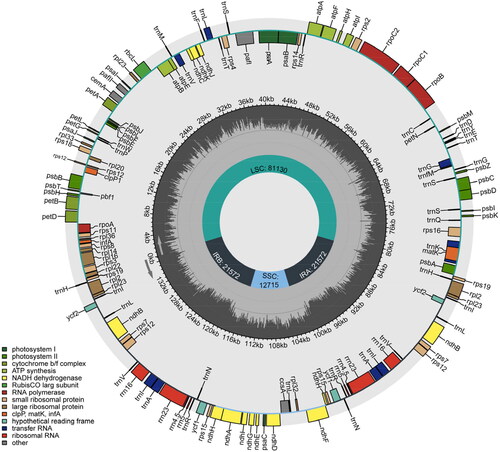
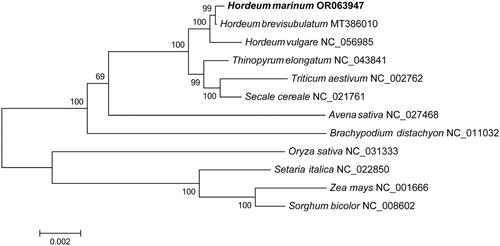
Figure 3. Phylogenetic tree inferred by ML method based on the complete chloroplast genome of 12 species. The complete chloroplast genomes of 11 species, including Hordeum brevisubulatum (MT386010) (Su et al. Citation2020), Hordeum vulgare (NC_056985), thinopyrum elongatum (NC_043841), Triticum aestivum (NC_002762) (Ogihara et al. Citation2002), secale cereale (NC_021761) (Middleton et al. Citation2014), avena sativa (NC_027468) (Saarela et al. Citation2015), brachypodium distachyon (NC_011032) (Bortiri et al. Citation2008), Oryza sativa (NC_031333), setaria italica (NC_022850), Zea mays (NC_001666) (Maier et al. Citation1995) and Sorghum bicolor (NC_008602) (Saski et al. Citation2007) were downloaded from NCBI (www.ncbi.nlm.nih.gov/).
Supplemental Material
Download MS Word (4.2 MB)Data availability statement
The chloroplast genome sequence data supporting the findings of this study are publicly available in NCBI. The complete chloroplast genome of H. marinum ssp. marinum was submitted to GenBank with the accession number OR063947. The associated BioProject is PRJNA976964; the SRA number is SRR24757234; and the Bio-Sample number is SAMN35447610, respectively.
