Figures & data
Figure 1. The photo of an individual of B. purpurascens was taken by Li Ao from Aba Tibetan and Qiang Autonomous Prefecture in Sichuan Province, China, including young leaves and flowers, the core feature of the species including its thick, large, and leathery of obovate leaves, cymose paniculate inflorescences, and broadly ovate purplish-red petals.

Figure 2. Graphic representation of features identified in the cp genome of B. purpurascens. Genes inside the circle are transcribed clockwise, while those outside are transcribed counterclockwise. Genes are color-coded according to functional groups. The dark pink region inside the inner circle indicates the GC content, while the green color indicates the at content of the cp genome. Boundaries of the small single copy (SSC) and large single copy (LSC) regions and the inverted repeat (IRa and IRb) regions are denoted in the inner circle.
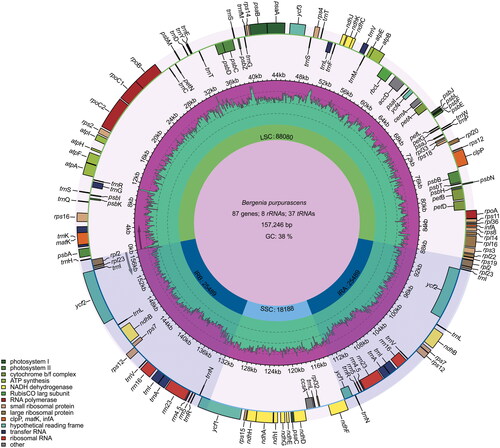
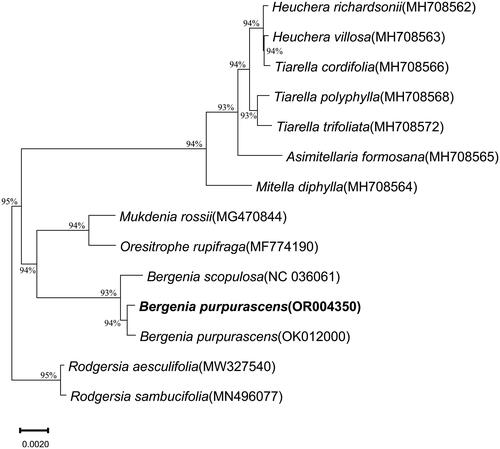
Figure 3. Phylogenetic relationships of Bergenia purpurascens inferred from maximum likelihood (ML) analysis. Numbers beside the node indicate bootstrap support values. The following sequences were used: B. purpurascens OR004350 (this study), OK012000 (Chen et al. Citation2022), B. scopulosa NC036061 (Bai et al. Citation2017), Heuchera richardsonii R.Br. 1823 MH708562, Heuchera villosa Michx. 1803 MH708563, Tiarella cordifolia L. 1753 MH708566, Tiarella polyphylla D. Don. 1825 MH708568, Tiarella trifoliata L. 1753 MH708572, Asimitellaria formosana R.A. Folk & Y. Okuyama. 2021 MH708565, Mitella diphylla L. 1753 MH708564, Mukdenia rossii Koidz. 1935 MG470844, Oresitrophe rupifraga Bunge 1835 MF774190, R. aesculifolia MW327540, R. sambucifolia MN496077 (Yang et al. Citation2022).
Supplemental Material
Download TIFF Image (4.7 MB)Supplemental Material
Download TIFF Image (102.6 MB)Supplemental Material
Download JPEG Image (1.2 MB)Data availability statement
The chloroplast genome sequence data in this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ with the accession number OR004350. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1004063, SRR25609650, and SAMN36922130, respectively.
