Figures & data
Figure 1. Photos of Chrysoglossum ornatum. (A) Inflorescences; (B) plants; (C) flower. The photographs were taken by Wei Wu.

Figure 2. The chloroplast genome map of Chrysoglossum ornatum. From the center outward, the map consists of six rings. The first circle represents the forward and reverse repeats connected with red and green arcs, respectively. The second circle shows the tandem repeats marked. The third circle displays the microsatellite sequences. The fourth circle indicates the sizes of feature regions, including a large single-copy (LSC), a small single-copy (SSC), and two inverted repeats (IRa and IRb). The fifth circle exhibits the GC content. The sixth circle presents the genes with different functions.
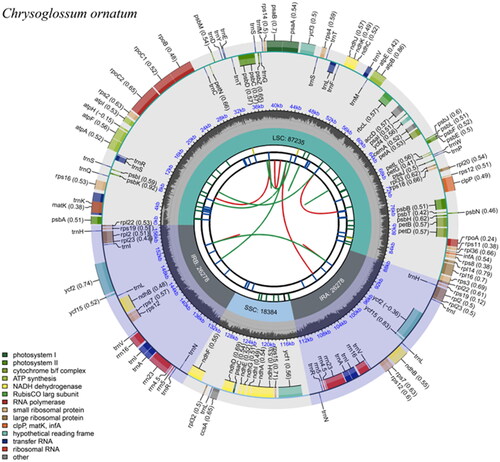
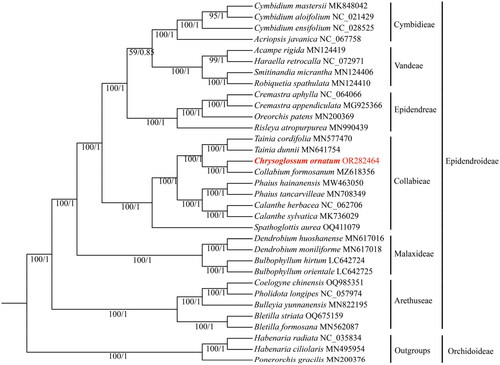
Figure 3. Phylogenetic tree inferred from the complete chloroplast genomes comprising 33 species from Orchidaceae. The ML bootstrap (BS) and BI posterior probability (PP) that supported each node are shown under the branches. The following sequences were used: Acampe rigida (Liu et al. Citation2020), Bletilla formosana (Wu et al. Citation2019), Bletilla striata (Feng et al. Citation2019), Bulbophyllum hirtum (Yang et al. Citation2022), Bulbophyllum Orientale (Yang et al. Citation2022), Bulleyia yunnanensis (Ai et al. Citation2020), Calanthe sylvatica (Miao et al. Citation2019), Cremastra appendiculata (Mao et al. Citation2018), Cymbidium aloifolium (Chen et al. Citation2020), Cymbidium ensifolium (Jiang et al. Citation2019), Cymbidium mastersii (Zheng et al. Citation2019), Habenaria ciliolaris (Chen et al. Citation2019), Oreorchis patens (Kim et al. Citation2020), Ponerorchis gracilis (Kim et al. Citation2020), Robiquetia spathulata (Liu et al. Citation2020), Smitinandia micrantha (Liu et al. Citation2020), Tainia cordifolia (Zheng et al. Citation2019), and Tainia dunnii (Xie et al. Citation2019).
Supplemental Material
Download MS Word (1.4 MB)Data availability statement
The chloroplast genome of Chrysoglossum ornatum was deposited in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OR282464. The associated BioProject, BioSample, and SRA numbers are PRJNA995913, SAMN36510063, and SRR25319442, respectively.
