Figures & data
Figure 1. Tricyrtis xianjuensis Li, Chen & Ma 2014. (A) Habitat; (B) seedling; (C) flower. All the photos were taken by Ming Jiang. Tricyrtis xianjuensis is a perennial herb up to 70 cm, with short rhizomes, ascending stems, alternate leaves, and yellow flowers. Flowering period of this species occurs between September and early October, while fruiting in October.

Figure 2. The chloroplast genome map of Tricyrtis xianjuensis. The map encompasses six tracks that depict various features of the genome. Starting from the center, the first track displays dispersed repeats, consisting of both direct and palindromic repeats, which are delineated by red and green arcs. The second track highlights long tandem repeats represented by short blue bars. In the third track, short tandem repeats or microsatellite sequences are illustrated as colored bars. Each color corresponds to a specific type of repeat, and accompanying descriptions provide valuable information about the characteristics of each repeat type. The colors and their respective repeat types are as follows: black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The chloroplast genome contains an LSC region, an SSC region, and two IR regions, and they are shown on the fourth track. The GC content along the genome is shown on the fifth track. Genes within the genome visualization are meticulously color-coded based on their functional classification. The transcription directions of the inner genes are represented in a clockwise manner, while the outer genes are shown in an anticlockwise orientation. To assist with interpretation, the key for gene functional classification is provided in the bottom left corner of the visualization.
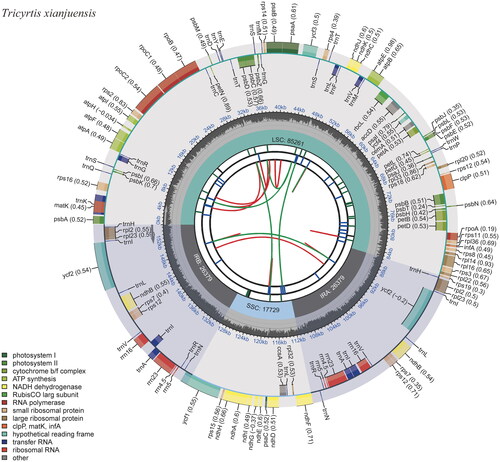
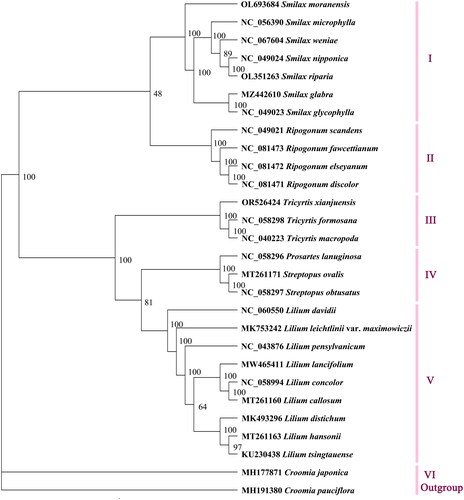
Figure 3. The maximum-likelihood tree based on complete chloroplast genome sequences of Tricyrtis xianjuensis and 25 species of family Liliaceae, with Croomia japonica and C. pauciflora as the outgroup species. The numbers next to the nodes are bootstrap support values. NCBI accession numbers of each genome are shown in the figure. The following sequences were used: Croomia pauciflora MH191380 (Lu et al. Citation2018), C. japonica MH177871 (Lu et al. Citation2018), Smilax moranensis OL693684 (Ji et al. Citation2022), S. microphylla NC_056390 (Wu et al. Citation2021), S. weniae NC_067604 (Feng et al. Citation2022), S. nipponica NC_049024, S. riparia OL351263, S. glabra MZ442610, S. glycophylla NC_049023 (Do et al. Citation2020), Ripogonum scandens NC_049021 (Do et al. Citation2020), R. fawcettianum NC_081473, R. elseyanum NC_081472, R. discolor NC_081471, Tricyrtis formosana NC_058298, T. macropoda NC_040223 (Wang et al. Citation2018), Prosartes lanuginose NC_058296, Streptopus ovalis MT261171 (Do et al. Citation2020), S. obtusatus NC_058297, Lilium davidii NC_060550 (Li et al. Citation2021), L. leichtlinii var. maximowiczii MK753242, L. pensylvanicum NC_043876 (Ramekar et al. Citation2019), L. lancifolium MW465411, L. concolor NC_058994, L. callosum MT261160 (Do et al. Citation2020), L. distichum MK493296, L. hansonii MT261163 (Do et al. Citation2020), and L. tsingtauense KU230438 (Song et al. Citation2016).
Supplemental Material
Download MS Word (351.8 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ OR526424. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1025334, SRR26334644, and SAMN37721183, respectively.
