Figures & data
Figure 1. Algal characteristics of V. punctata. Panel A presents the morphological observations of cells under optical microscopy. The cells appear globular or angular, some with small humps, single parietal, much-lobed chloroplast and a single protruded polyhedral pyrenoid. Mature vegetative cells often reveal a cytoplasmic lipidic reddish globule. Panel B portrays V. punctata cultivated in bubble column glass photobioreactors. Panel C illustrates the color change of cells over time, from yellow-green on day one to dark green on day six. Panel D demonstrates V. punctata cells harvested on day six, which were used for DNA extraction in this study. These images were captured by Luodong Huang on 12 September 2020, at the Biological Resources and Environmental Microbiology Laboratory of the College of Life Science and Technology, Guangxi University.
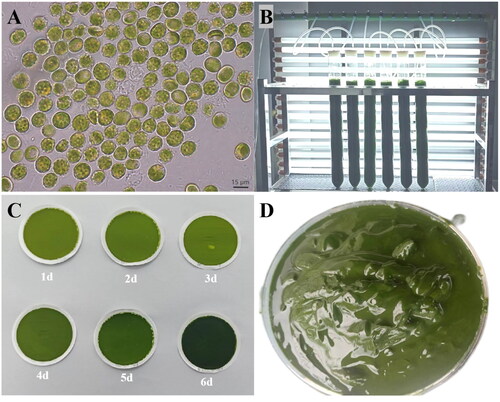
Figure 2. The circular map of the V. punctata mitochondrial genome. Genes belonging to different functional groups are color coded. Differences in colors lumps represent different gene types.
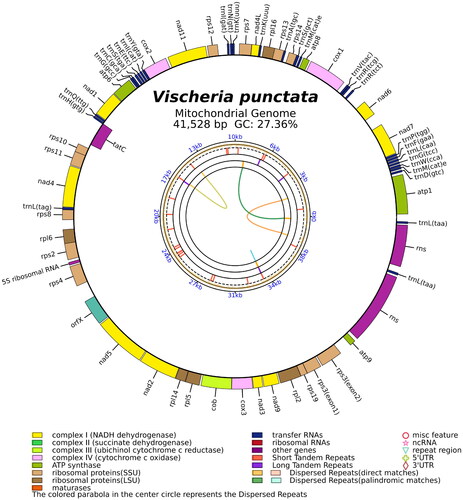
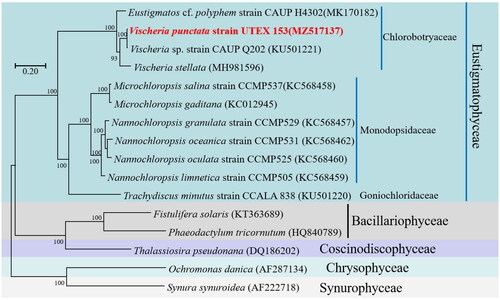
Figure 3. Phylogenetic relationships among 16 Ochrophyta mitochondrial genomes. Node labels indicate bootstrap values. The scale bar corresponds to 0.20 substitutions per nucleotide position. GenBank accession numbers for the utilized sequences are indicated within parentheses. Eustigmatos cf polyphem (MK170182) (Huang et al. 2018#), Vischeria sp. CAUP Q202 (KU501221) (Ševčíková et al. Citation2016), Vischeria stellata (MH981596) (Huang et al. Citation2019), Microchloropsis salina (KC568458) (Wei et al. Citation2013), Microchloropsis gaditana (KC012945), Nannochloropsis granulata (KC568457) (Wei et al. Citation2013), Nannochloropsis oceanica (KC568462) (Wei et al. Citation2013), Nannochloropsis oculata (KC568460) (Wei et al. Citation2013), Nannochloropsis limnetica (KC568459) (Wei et al. Citation2013), Trachydiscus minutus (KU501220) (Ševčíková et al. Citation2016), Fistulifera solaris (KT363689) (Tang and Bi Citation2016), Phaeodactylum tricornutum (HQ840789) (Oudot-Le and Green Citation2011), Thalassiosira pseudonana (DQ186202) (Armbrust et al. Citation2004), Ochromonas danica (AF287134) (Burger et al. 2016#), Synura synuroidea (AF222718) (Chesnick et al. Citation2000). #Direct submission to NCBI, unpublished.
Supplemental Material
Download PDF (96.4 KB)Supplemental Material
Download MS Word (157.5 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under accession no. MZ517137.1. The associated BioProject, BioSample, and SRA numbers are PRJNA759399, SAMN21166780, and SRR15691269, respectively.
