Figures & data
Figure 1. The reference image of Pinus bhutanica living in Linzhi, Xizang, China (geographic coordinates: 29°13′27″ N, 95°11′3″ E; photography by Yixuan Kou and Jing Wang).

Figure 2. Genomic map of overall features of Pinus bhutanica chloroplast genome, generated by CPGView (Liu et al. Citation2023). The map contains six tracks by default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct and palindromic repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The genome length and the GC content along the genome are shown on the fourth and fifth track, respectively. The genes are shown on the sixth track. Genes are color-coded by their functional classification.
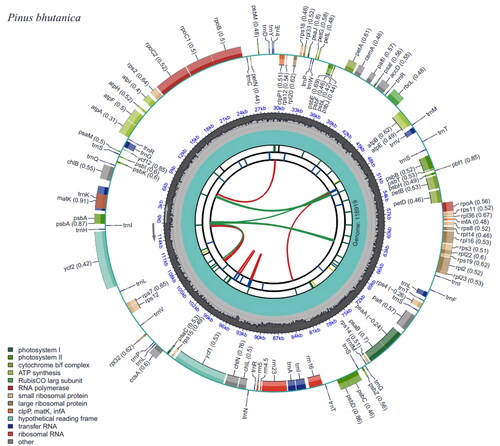
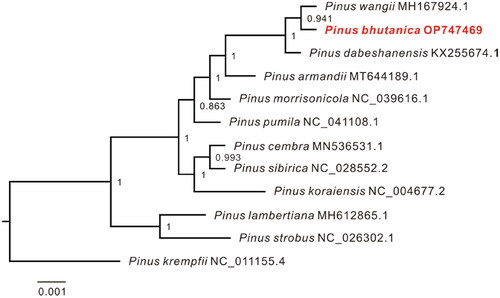
Figure 3. Maximum-likelihood (ML) tree inferred by FastTree with the GTR substitution model based on the complete chloroplast genome sequences of P. bhutanica and 10 other species in subsection Strobus. The numbers on the nodes represent the support values. The following sequences were used: Pinus wangii MH167924.1 (Yang et al. Citation2018), Pinus dabeshanensis KX255674.1 (Duan et al. Citation2016), Pinus armandii MT644189.1 (Jia et al. Citation2020), Pinus morrisonicola NC_039616.1 (Zeb et al. Citation2020), Pinus pumila NC_041108.1 (Zeb et al. Citation2019), Pinus cembra MN536531.1 (Schott et al. Citation2019), Pinus sibirica NC_028552.2 (Baturina et al. Citation2019), Pinus koraiensis NC_004677.2 (available in the GenBank of NCBI), Pinus lambertiana MH612865.1 (Gernandt et al. Citation2018), Pinus strobus NC_026302.1 (Zhu et al. Citation2016), and Pinus krempfii NC_011155.4 (Cronn et al. Citation2008).
Supplemental Material
Download PDF (3.3 MB)Supplemental Material
Download PDF (33.1 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OP747469. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA898140, SRR22188315, and SAMN31601598, respectively. A specimen of Pinus bhutanica was deposited at Jiangxi Agricultural University (https://www.jxau.edu.cn; contact person and email: Jing Wang, [email protected]) under the voucher number 2021-PB-1A.
