Figures & data
Figure 1. Reference image of M. corchorifolia. This image was taken by Wen Wang in the Baicao Garden of Zhejiang Chinese Medical University. The plant height is 1–6 ft; branches are yellowish-brown; leaves are variable in shape, simple, petiolate, ovate to lanceolate in outline, and doubly serrate. Simple hairs occur along the major veins on the lower surface of the leaf; the leaf base is round or heart-shaped, and stipules are bar-shaped; flowers are produced in terminal head-like cymes and are arranged in umbrella inflorescences or clusters of umbrella inflorescences apically or axillarily, with petals that can change from white to reddish.

Figure 2. Map of the circular chloroplast genome of M. corchorifolia. Various functional genes were annotated across the assembled genome, as indicated by different colors. Genes outside the circle were transcribed in a counterclockwise direction, whereas genes inside the circle were transcribed in a clockwise direction. The circular chloroplast genome consisted of the inverted repeats, the small single-copy, and the large single-copy sequences. From the outside toward the Central grey inner circle, dark grey represents the GC content, and light grey represents the at content.
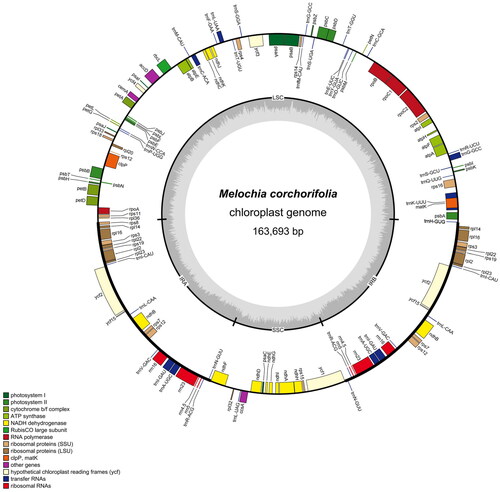
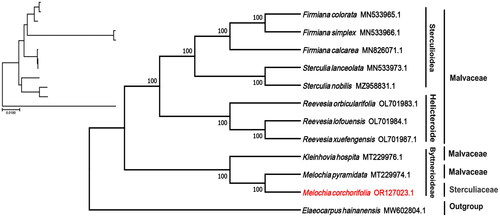
Figure 3. Phylogenetic tree of the complete chloroplast genome of M. corchorifolia with bootstrap values at each node. Sequences of the other species used to make the phylogenetic tree: Melochia corchorifolia (OR127023.1; this study), Melochia pyramidata (MT229974.1; unpublished), Kleinhovia hospita (MT229976.1; unpublished), Reevesia orbicularifolia (OL701983.1; unpublished), Reevesia lofouensis (OL701984.1; unpublished), Reevesia xuefengensis (OL701987.1; unpublished), Firmiana calcarea (MN826071.1; Lu and Luo Citation2022), Firmiana colorata (MN533965.1; Lu and Luo Citation2022), Firmiana simplex (MN533966.1; Lu and Luo Citation2022), Sterculia lanceolata (MN533973.1; unpublished), and Sterculia nobilis MZ958831.1; Lu and Luo Citation2022); Elaeocarpus hainanensis (MW602804.1; unpublished) was used as outgroup.
Supplemental Material
Download MS Word (716.5 KB)Supplemental Material
Download PDF (69.1 KB)Data availability statement
The sequence information of the complete chloroplast genome assembly of M. corchorifolia is available on the NCBI website (https://www.ncbi.nlm.nih.gov/) with the accession number OR127023.1. BioProject, Bio-Sample, and SRA are PRJNA1000299, SAMN36767426, and SRR25460835, respectively.
