Figures & data
Figure 1. Photographs of (A) flowering branches and (B) capitulum of Duhaldea cappa. Duhaldea cappa is a shrub, 70–200 cm tall; stems lanate-tomentose, branched; leaf blade elliptic, lanceolate, or narrowly oblong; capitula radiate or disciform, in dense corymbs. Photos were taken by Dr. Zhixi Fu in Chuxiong city, Yunnan province, China (101°29′41.28″E, 25°1′33.59″N), August 2020 without any copyright issues.

Figure 2. Genomic map of overall features of Duhaldea cappa chloroplast genome, generated by CPGview. The species’ name is shown in the top left corner. The map contains six tracks by default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
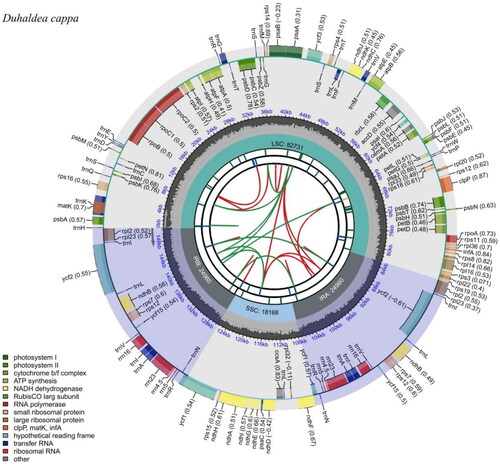
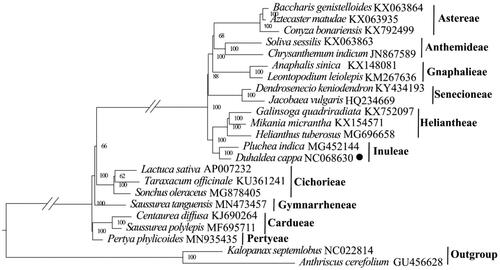
Figure 3. Maximum-likelihood phylogeny of Duhaldea cappa and related taxa based on 23 complete chloroplast genomes. The numbers on the branches represent the bootstrap values based on 1000 replicates. The sequences used for tree construction are as follows: Baccharis genistelloides (KX063864; Vargas et al. Citation2017), Aztecaster matudae (KX063935; Vargas et al. Citation2017), Conyza bonariensis (KX792499; Wang et al. Citation2018), Leontopodium leiolepis (KM267636), Anaphalis sinica (KX148081), Chrysanthemum indicum (JN867589), Soliva sessilis (KX063863; Vargas et al. Citation2017), Jacobaea vulgaris (HQ234669; Doorduin et al. Citation2011), Dendrosenecio keniodendron (KY434193), Galinsoga quadriradiata (KX752097; Wang et al. Citation2018), Mikania micrantha (KX154571; Huang et al. Citation2016), Helianthus tuberosus (MG696658), Pluchea indica (MG452144; Zhang et al. Citation2017), Lactuca sativa (AP007232), Taraxacum officinale (KU361241), Sonchus oleraceus (MG878405; Hereward et al. Citation2018), Saussurea tanguensis (MN573457), Saussurea polylepis (MF695711; Yun et al. Citation2017), Centaurea diffusa (KJ690264), Pertya phylicoides (MN935435; Wang et al. Citation2020), Anthriscus cerefolium (GU456628; Downie and Jansen Citation2015), Kalopanax septemlobus (NC022814; Li et al. Citation2013). The circle represents newly sequenced species (Duhaldea cappa genbank No. NC068630).
Supplemental Material
Download MS Word (315.4 KB)Data availability statement
After uploading the data, the NCBI database releases two accession numbers of Duhaldea cappa. These two accession numbers contain exactly the same sequence information and author. Data are available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov (accession number: NC068630 and OM457000). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA938187, SRR23606895 and SAMN33427715, respectively.
