Figures & data
Figure 1. Photograph of Asplenium pseudocapillipes taken by Sang Hee Park at the collection site. This plant is epilithic. Its fronds are cespitose, herbaceous, and subglabrous. Furthermore, gemma is not present on the fronds.

Figure 2. Complete chloroplast genome map of A. pseudocapillipes, containing six tracks. From the center, the first track shows the dispersed repeats that have direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, containing a large single copy, a small single copy, and two inverted regions, are identified on the fourth track. The guanine–cytosine content along the genome is plotted in the fifth track. The genes are shown on the outer sixth track.
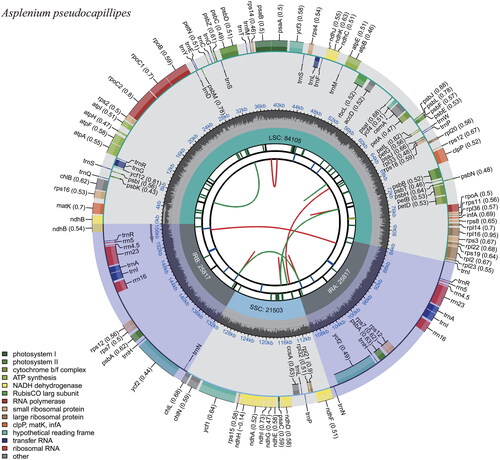
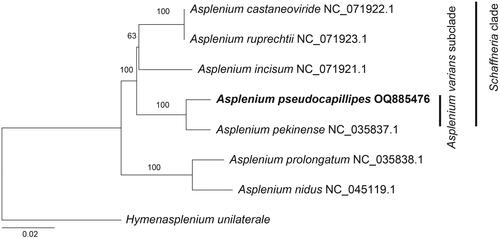
Figure 3. A maximum likelihood tree was constructed based on 83 cp coding genes, with bootstrap support values indicated on the nodes. The phylogenetic position of Asplenium pseudocapillipes was investigated by including six other Asplenium species (A. castaneoviride, NC_071922; A. incisum, NC_071921; A. nidus, NC_045119; A. pekinense, NC_035837; A. prolongatum, NC_035838; and A. ruprechtii, NC_071923) in the analysis. The outgroup taxon was Hymenasplenium unilaterale (NC_035856).
Supplemental Material
Download MS Word (16.7 MB)Supplemental Material
Download EPS Image (1,016.4 KB)Supplemental Material
Download EPS Image (9.2 MB)Supplemental Material
Download MS Word (15 MB)Data availability statement
The genome sequence data supporting the findings of this study are available in NCBI GenBank under accession number OQ885476. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA962509, SRR24364397, and SAMN34403414, respectively.
