Figures & data
Figure 1. Appearance of Prunus salicina cultivar ‘Zuili’. The photo was taken by Liangling Cao at Tongxiang, Jiaxing city, Zhejiang Province, China (30°42′38″N and 120°31′38″E). The fruit weighs around 50 g and has a dark purple-red skin adorned with a patchwork of varying yellow-brown spots. The leaves are simple, ovate to oblong in shape, with a finely serrated margin.

Figure 2. Chloroplast genome map of Prunus salicina cultivar ‘Zuili’ by CPGview. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows: black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The quadripartite structure (LSC, SSC, IRA, and IRB) is illustrated on the fourth track. The GC content is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
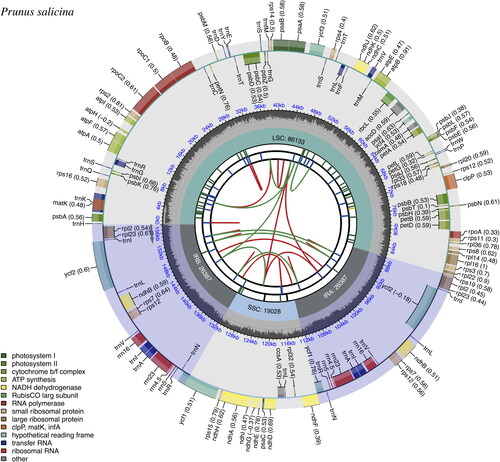
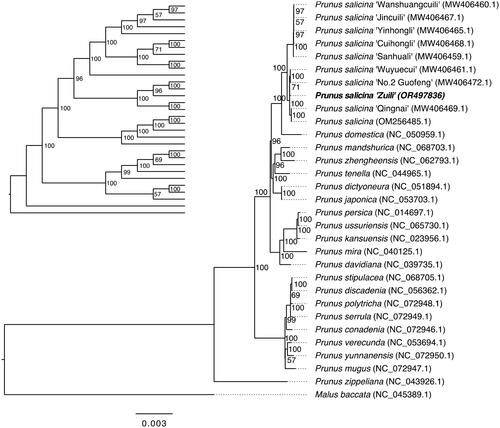
Figure 3. Phylogenetic tree inferred by maximum-likelihood (ML) method based on the complete chloroplast genomes of 30 Prunus species and one outlier (Malus baccata). For clearly displaying the phylogenetic relationship, a cladogram without species name is placed in the top left. A total of 1000 bootstrap replicates were performed, and the bootstrap support values (percentage) are indicated behind nodes. The chloroplast genome of Prunus salicina cultivar Zuili is shown in bold. The NCBI GenBank accession numbers are shown in parentheses. The gray lines were used to supplement the branch length, and the branch length of the phylogram was black. The following sequences were used: Prunus salicina ‘Wanshuangcuili’ MW406460.1, Prunus salicina ‘Jincuili’ MW406467.1, Prunus salicina ‘Yinhongli’ MW406465.1, Prunus salicina ‘Cuihongli’ MW406468.1, Prunus salicina ‘Sanhuali’ MW406459.1, Prunus salicina ‘Wuyuecui’ MW406461.1, Prunus salicina ‘No.2 Guofeng’ MW406472.1, Prunus salicina ‘Qingnai MW406469.1, Prunus salicina OM256485.1 (Su et al. Citation2023), Prunus domestica NC_050959.1 (Geng et al. Citation2020), Prunus mandshurica NC_068703.1, Prunus zhengheensis NC_062793.1, Prunus tenella NC_044965.1, Prunus dictyoneura NC_051894.1 (Liu et al. Citation2019), Prunus japonica NC_053703.1 (Mu et al. Citation2021), Prunus persica NC_014697.1 (Jansen et al. Citation2011), Prunus ussuriensis NC_065730.1, Prunus kansuensis NC_023956.1, Prunus mira NC_040125.1 (Bao et al. Citation2019), Prunus davidiana NC_039735.1 (Zhang et al. Citation2018), Prunus stipulacea NC_068705.1, Prunus discadenia NC_056362.1, Prunus polytricha NC_072948.1, Prunus serrula NC_072949.1, Prunus conadenia NC_072946.1, Prunus verecunda NC_053694.1 (Jiang et al. Citation2019), Prunus yunnanensis NC_072950.1, Prunus mugus NC_072947.1, Prunus zippeliana NC_043926.1, and Malus baccata NC_045389.1.
Supplemental Material
Download MS Word (794.7 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OR497836.1. The associated ‘BioProject’, ‘SRA’, and ‘Bio-Sample’ numbers are PRJNA1026316, SRR26329588, and SAMN37737007, respectively.
