Figures & data
Figure 1. Morphology of QHP. The photo was taken by Weibing Zhuang at Nanjing Botanical Garden, Memorial Sun Yat-sen (E118_83, N32_06), Nanjing, China, on 24 June 2020, without any copyright issues. Leaves and new shoots of QHP were dark purple from bud flush in spring to late June, then showing a medium shade of purple from July to September, and turning into bright red in October. The bright and reddish-purple leaves among ‘QHP’ are the main characters distinct from other poplar cultivars.

Figure 2. The chloroplast genome map of QHP. Genes shown outside the circle are transcribed clockwise, and genes inside are transcribed counter-clock-wise. Genes belonging to different functional groups are color-coded. The darker grey in the inner corresponds to the GC content and the lighter grey to the AT content.
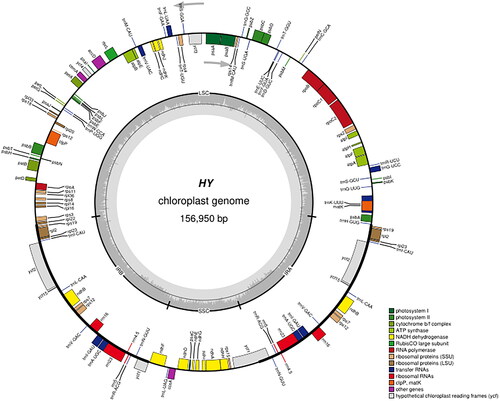
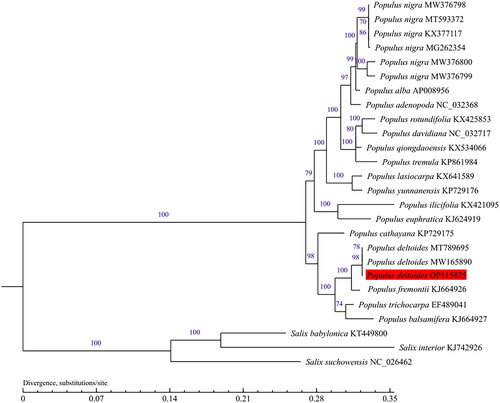
Figure 3. Phylogenetic tree was built with maximum-likelihood (ML) bootstrap analysis based on 23 chloroplast genome sequences from Populus and three taxa from Salix were served as outgroups. Label annotations against each species name represent the accession number on GenBank. The numbers above branches indicate bootstrap support values (%) of neighbor joining maximum likelihood trees, respectively. The bootstrap values are evaluated based on 1000 replications. The scale bar represents the number of substitutions at each locus. The following sequences were used: Populus nigra KX377117, Populus cathayana KP729175, Populus tremula KP861984, Populus Alba AP008956, Populus yunnanensis KP729176 (He et al. Citation2019), Salix suchowensis NC_026462, Populus adenopoda NC_032368 (Wu et al. Citation2020), Populus balsamifera KJ664927, Populus euphratica KJ624919, Populus fremontii KJ664926, Populus ilicifolia KX421095, Populus lasiocarpa KX641589, Populus qiongdaoensis KX534066, Populus rotundifolia KX425853, Populus trichocarpa EF489041 (Zhang et al. Citation2019), Populus rotundifolia KX425853, Salix interior KJ742926 (Zong et al. Citation2019), Populus deltoides MT789695, Populus davidiana NC_032717, Salix babylonica KT449800 (Zhuang et al. Citation2020), Populus deltoides MW165890 (Zhuang et al. Citation2021), Populus nigra MW376800, Populus nigra MW376799, Populus nigra MW376798 (Wang et al. Citation2022), and Populus nigra MG262354, Populus nigra MT593372 (unpublished).
Supplemental Material
Download MS Excel (57 KB)Supplemental Material
Download MS Word (1.3 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OP115875. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA 862560, SRR16145711, and SAMN29984011, respectively.
