Figures & data
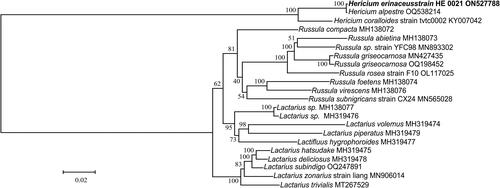
Figure 3. The maximum-likelihood (ML) phylogenetic position of H. erinaceus based on the 15 common protein-coding genes in each complete mitogenome sequence from 22 species. The values on the branches are the ML bootstrap percentages. The mitogenome of H. erinaceus determined in this study is marked by bold-type. Scale bar is for substitutions per site. The following sequences were used: Russula subnigricans, R. virescens, R. abietina, R. compacta, and R. foetens (Yu et al. Citation2019), R. griseocarnosa, R. rosea, Lactarius deliciosus, and L. hatsudake (Yu and Liang Citation2022), L. trivialis (Shao et al. Citation2020), L. hygrophoroides (Cai et al. Citation2021), L. volemus (Sun et al. Citation2020), H. coralloides (Zhang et al. Citation2017), and H. alpestre, Russula sp., L. zonarius, Lactarius sp., L. subindigo, and L. piperatus (unpublished).
Supplemental Material
Download PDF (256.1 KB)Data availability statement
The genome sequence data supporting the findings of this study are available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. ON527788. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA869545, SRR21047616, and SAMN30309510, respectively.