Figures & data
Figure 1. Underwater image of the Hyperhalosydna striata. The photo was taken by TP, a corresponding author of this paper.

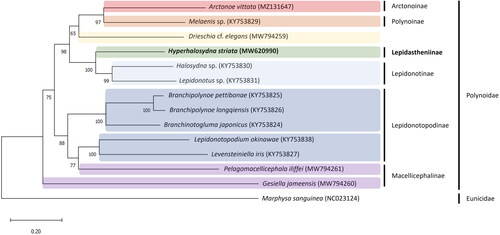
Figure 3. Maximum-likelihood (ML) tree reconstructed using a concatenated data set of 13 protein-coding genes based on 14 mitogenome sequences, including Hyperhalosydna striata from the present study. Bootstrap replicates were performed 1000 times. The GenBank accession number of each species is shown in parentheses after the species name. The following sequences were used: Hyperhalosydna striata MW620990, Arctonoe vittata MZ131647 (Park et al. Citation2021), Melaenis sp. KY753829 (Zhang et al. Citation2018), Drieschia cf. elegans MW794259 (Gonzalez et al. Citation2021), Halosydna sp. KY753830 (Zhang et al. Citation2018), Lepidonotus sp. KY753831 (Zhang et al. Citation2018), Branchipolynoe pettibonae KY753825 (Zhang et al. Citation2018), B. longqiensis KY753826 (Zhang et al. Citation2018), Branchinotogluma japonicus KY753824 (Zhang et al. Citation2018), Lepidonotopodium okinawae KY753838 (Zhang et al. Citation2018), Levensteiniella iris KY753827 (Zhang et al. Citation2018), Pelagomacellicephala iliffei MW794261 (Gonzalez et al. Citation2021), and Gesiella jameensis MW794260 (Gonzalez et al. Citation2021).
Supplemental Material
Download PNG Image (45.6 KB)Data availability statement
The genome sequence data supporting this study’s findings are available in the National Center for Biotechnology Information (NCBI) GenBank (https://www.ncbi.nlm.nih.gov) under accession no. MW620990. The associated BioProject, SRA, and BioSample numbers were PRJNA727906, SRR14566003, and SAMN19229862, respectively. The data that support the findings of this study are openly available in Mendeley (https://doi.org/10.17632/9h4gtkx9fn.1).