Figures & data
Figure 1. Species reference map (Fruits and leaves) of S. fulvum (Wild species; Voucher number: ZM02301; This picture was taken by Guangbo Zhang from the Yunnan Agricultural University, Yunnan Province, China; 102.75574°E, 25.13488°N). S. fulvum is a near-source wild species of sugarcane that has the advantages of early maturation, high sugar content, and drought resistance that is of great value to the genetic improvements of sugarcane varieties. Core features: Culms erect, nodes long-mustachioed, upper and lower parts of nodes pilose or white powdery. Leaf blades long linear and ciliate. Panicle. Spikelet sessile and lanceolate, with white filiform hairs. Chromosome number 2n = 20. Flowering and fruiting period August-November.

Figure 2. The chloroplast genome map of S. fulvum. Genes on the inside of the circle are transcribed in a clockwise direction and genes on the outside of the circle are transcribed in a counter-clockwise direction.
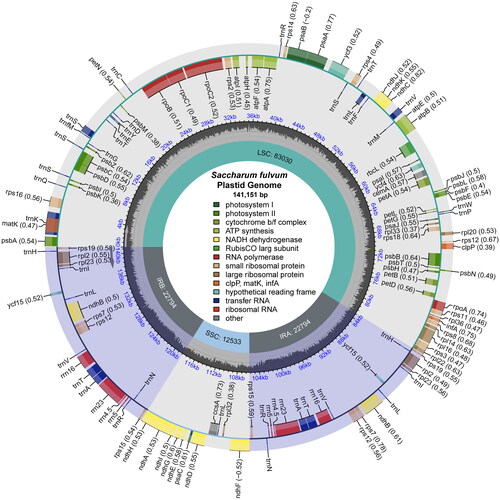
Table 1. Summary of the chloroplast genomes of S. fulvum and S. narenga species.
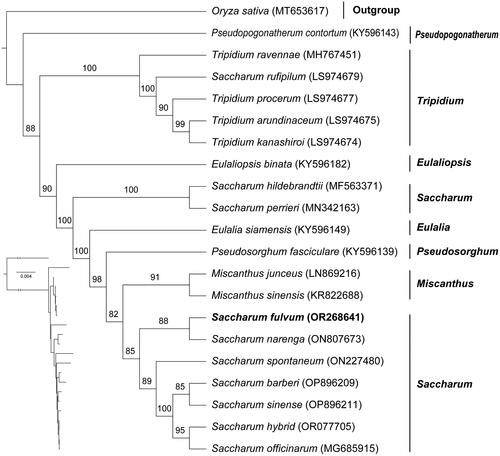
Figure 3. Phylogenetic tree based on the concatenated sequences of 76 protein-coding genes in 21 species by maximum-likelihood (ML). Values split by backslashes above branches represent ML bootstraps. The best-fit model was TPM3 + F + R5. Branch supports were tested using ultrafast bootstrap with 1000 replicates. Saccharum fulvum (OR268641) was marked in bold. The following sequences were used: Oryza sativa (voucher: HSAGSDYD1802) (MT653617) (Fan et al. Citation2020), Pseudopogonatherum contortum (KY596143), Tripidium ravennae (MH767451) (Lloyd Evans et al. Citation2019), Saccharum rufipilum (LS974679 (Lloyd Evans et al. Citation2019), Tripidium procerum (LS974677) (Lloyd Evans et al. Citation2019), Tripidium arundinaceum (LS974675) (Lloyd Evans et al. Citation2019), Tripidium kanashiroi (LS974674) (Lloyd Evans et al. Citation2019), Eulaliopsis binata (KY596182), Saccharum hildebrandtii (MF563371) (Piot et al. Citation2018), Saccharum perrieri (MN342163), Eulalia siamensis (KY596149), Pseudosorghum fasciculare (KY596139), Miscanthus junceus (LN869216), Miscanthus sinensis (KR822688), Saccharum narenga (ON807673) (Dyfed Lloyd and Ben, Citation2020), Saccharum spontaneum (ON227480) (Evans and Joshi, Citation2016), Saccharum barberi (OP896209) (Li et al. Citation2022), Saccharum sinense (OP896211) (Li et al. Citation2022), Saccharum hybrid (OR077705) (Li et al. Citation2022), Saccharum officinarum (MG685915) (Evans and Joshi, Citation2016).
Supplemental Material
Download MS Excel (15.2 KB)Supplemental Material
Download MS Excel (14.6 KB)Supplemental Material
Download MS Excel (12 KB)Supplemental Material
Download JPEG Image (821 KB)Supplemental Material
Download JPEG Image (1.4 MB)Supplemental Material
Download JPEG Image (443.5 KB)Supplemental Material
Download JPEG Image (1.1 MB)Data availability statement
The genome sequence data that support the findings of this study are available in GenBank of NCBI (http://www.ncbi.nlm.nih.gov/) under the accession no. OR268641. The associated BioProject, BioSample, and SRA numbers are PRJNA1005584, SAMN36990507, SRR25653907, respectively.
