Figures & data
Figure 1. A photo of the humped rockcod Gobionotothen gibberifrons (Lönnberg 1905). The photo has been taken by Jin-Hyoung Kim.

Figure 2. Circular gene map of the Gobionotothen gibberifrons mitochondrial genome, created and visualized by the MitoFish webserver (Sato et al. Citation2018) and Chloroplot software (Zheng et al. Citation2020), respectively. The inner circle of the image represents the GC% per every 5 bp of the mitogenome, with darker lines indicating higher GC% levels. The arrows inside and outside the innermost circle indicate the transcription direction of genes encoded on the light and heavy strands, displayed inside and outside the outermost circle, respectively.
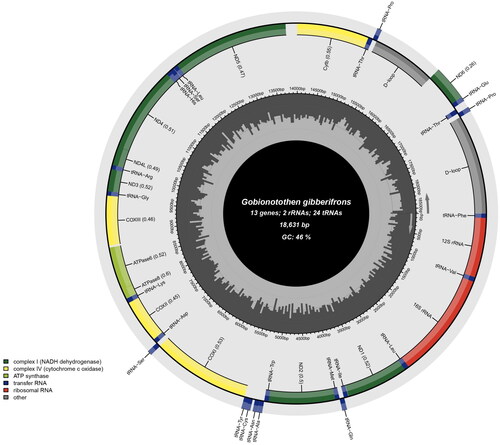
Figure 3. The maximum likelihood phylogenetic tree for Gobionotothen gibberifrons with Gadus morhua as outgroup. The tree was constructed using the amino acid sequences of 13 protein-coding genes. The following GenBank accession numbers of sequences were used: T. lepidorhinus MN864240; T. loennbergii MT447073 (Choi et al. Citation2021); T. nicolai MZ779013 (Patel et al. Citation2022); T. bernacchii MZ779012 (Patel et al. Citation2022); P. borchgrevinki MZ779011 (Patel et al. Citation2022); T. newnesi OP432732 (Nguyen et al. Citation2023); A. mitopteryx MT232658; D. eleginoides JAOVFM010000447; D. mawsoni LC138011; N. coriiceps JF933906 (Oh et al. Citation2016); N. rossii MN893242; P. antarcticum JF933905 (Lee et al. Citation2015); G. morhua X99772 (Johansen and Bakke Citation1996). The species in this study is highlighted by a black dot with bold font.

Supplemental Material
Download MS Word (632.9 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/under the accession no. OP804623. The associated BioProject, SRA, and BioSample numbers are PRJNA881762, SRR22406005, and SAMN31867186, respectively.
