Figures & data
Figure 1. Eremurus zoae Vved 1971 (Photographs was taken by Georgy A. Lazkov in Issyk Kul, Kyrgyz Republic). A, Habitat; B, Flower; C, Capsule. E. zoae is a perennial herb with a height of 25–40 cm. The flowers are campanulate, with yellow petals that reflex in fruiting. The capsules are globose. The flowering period is from April to May and the fruiting period is from May to June.

Figure 2. Complete chloroplast genome map of Eremurus zoae Vved. drawn using CPGview. The circle map contains six tracks. The first track indicates the repeat distribution. The second track shows the tandem repeats as a blue bar. The third track represents microsatellite sequences in green and yellow. The fourth track shows the LSC, SSC, and IR regions. The fifth track indicates the GC contents, and the sixth track represents genes.
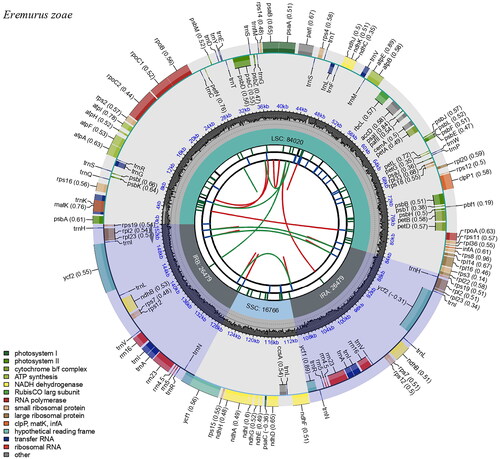
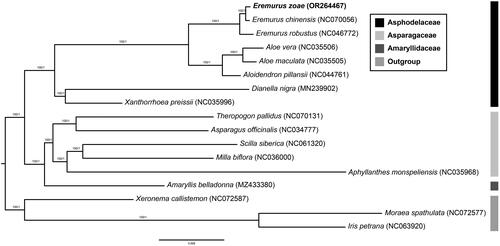
Figure 3. Phylogenetic tree of 17 species with three outgroup taxa (one Xeronemataceae, two Iridaceae) based on 82 protein-coding genes using the ML and BI methods. The numbers above the nodes indicate the bootstrap support values and the Bayesian posterior probabilities. The newly reported chloroplast genome in this study is indicated in red. All sequences used in the analysis are as follows: Eremurus zoae (OR264467, this study), Eremurus chinensis (NC070056), Eremurus robustus (NC046772, Makhmudjanov et al.Citation2019), Aloe vera (NC035506), Aloe maculata (NC035505), Aloidendron pillansii (NC044761), Dianella nigra (MN239902), Xanthorrhoea preissii (NC035996), Theropogon pallidus (NC070131), Asparagus officinalis (NC034777), Scilla siberica (NC061320), Milla biflora (NC036000), Aphyllanthes monspeliensis (NC035968), Amaryllis belladonna (MZ433380), Xeronema callistemon (NC072587, Kamra et al.Citation2023), Moraea spathulata (NC072577, Kamra et al.Citation2023), and Iris petrana (NC063920, Volis et al.Citation2022).
Supplemental Material
Download MS Word (481.7 KB)Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www. ncbi. nlm. nih. gov) under accession no. OR264467. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA996712, SRR25367042, and SAMN36621056, respectively.
