Figures & data
Figure 1. Three adult individuals of Rhynchophorus phoenicis (photo credit BJR; OnePlus 8, f/1.8, 1/1000). The species generally reaches body lengths, including the rostrum, of 3.5–4.5 cm (Tanyi Tambe et al. Citation2013).

Table 1. Publicly available mitochondrial genomes used in the phylogenetic analyses.
Figure 2. The mitochondrial genome of the African palm weevil (Rhynchophorus phoenicis). The 13 protein-coding genes, 22 tRNA, and two rRNA genes are shown in green, red, and orange, respectively. This plot was produced in MitoZ (Meng et al. Citation2019). GC content, in 50 bp windows, is shown by the blue bars, with the red ring that of 50% GC content, and genome-wide depth coverage is shown by the purple ring.
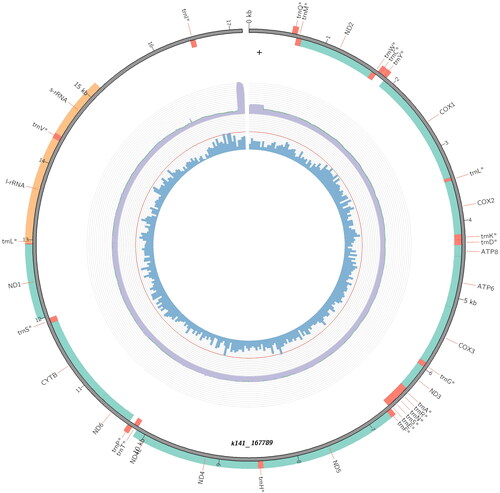
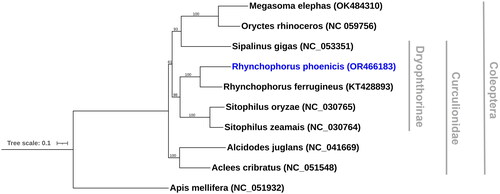
Figure 3. Maximum likelihood phylogenetic tree of nine coleopteran mitogenomes, including the new assembly produced here (in blue), and a single hymenopteran outgroup. Bootstrap support values, as a percentage of 1000 replicates, are given on nodes. GenBank accession numbers for publicly obtained sequences are shown in brackets and the papers they derive from are given in . Branch lengths are proportional to the number of substitutions per site (see scale bar).
Supplemental Material
Download MS Word (137.2 KB)Supplemental Material
Download JPEG Image (172.2 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OR466183. Associated BioProject, SRA, and BioSample numbers are PRJNA1037096, SRR26723342, and SAMN38162519, respectively.
