Figures & data
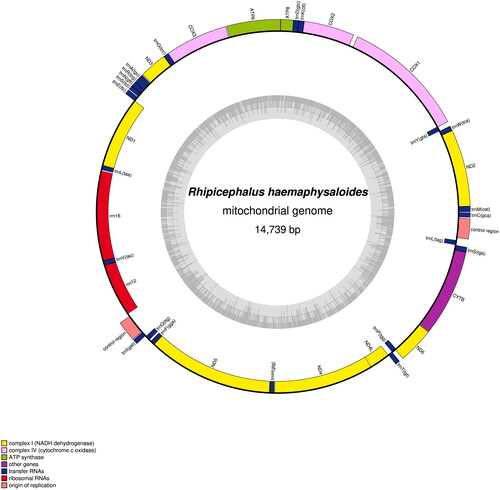
Table 1. Rhipicephalus haemaphysaloides gene content, length, coding strand, initiation, stop codons of mitochondrial genomes of different isolate.
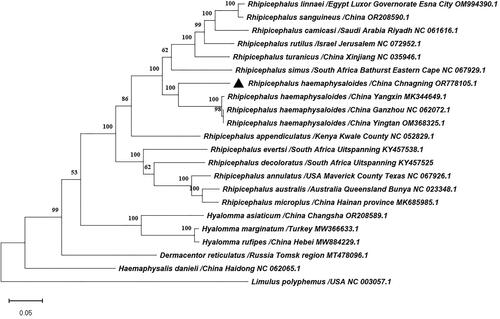
Figure 3. Phylogenetic tree of Rhipicephalus haemaphysaloides and 20 previously published ixodidae tick species in GenBank based on the nucleotides of 13 PCGs of mitochondrial genomes used the maximum likelihood method by MEGA11.0, the numbers at the nodes are bootstrap values computed using 1,000 replications and General Time Reversible model. The following sequences were used: Dermacentor reticulatus/Russia Tomsk region (Kartashov et al. Citation2020), Rhipicephalus australis/Australia Queensland, Bunya (Burger et al. Citation2014), Rhipicephalus camicasi/Saudi Arabia Riyadh (Chandra et al. Citation2022), Rhipicephalus decoloratus/South Africa Uitspanning (Mans et al. Citation2019), Rhipicephalus evertsi/South Africa Uitspanning (Mans et al. Citation2019), Rhipicephalus linnaei/Egypt Luxor Governorate, Esna City (Šlapeta et al. Citation2022), Rhipicephalus sanguineus/China (Cao et al. Citation2023), Hyalomma asiaticum/China Changsha (Cao et al. Citation2023), Hyalomma marginatum/Turkey (Ciloglu et al. Citation2021), Hyalomma rufipes/China Hebei (Lang et al. Citation2022), Limulus polyphemus/USA (Lavrov et al. Citation2000).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The accession number of the complete mitochondrial genome is OR778105. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA1054497, SRR27313610, and SAMN38923201, respectively.