Figures & data
Figure 1. The basidiocarps of Tylopilus brunneirubens collected from Jiangxi Province, China. The grayish red to brownish red or rust brown when bruised and its distinct reticulation on the upper stem are the most distinguished features. Photographed by Kuan Zhao.

Figure 2. The mitochondrial genome map of Tylopilus brunneirubens. Genes shown outside and inside the outer circle are transcribed in counterclockwise and clockwise directions, respectively. The inner circles represent the genome scale, GC content and distributions of short tandem repeats, long tandem repeats, and the dispersed repeats, respectively. The colored parabolas in the center circle represent the dispersed repeats.
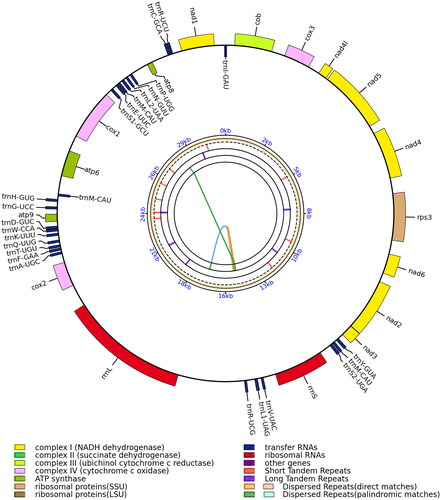
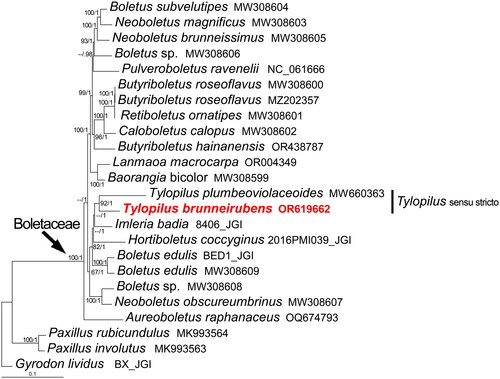
Figure 3. Phylogenetic tree of Tylopilus brunneirubens and related taxa based on Bayesian’s inference (BI) and maximum-likelihood (ML) analyses of 15 core protein coding genes (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and rps3). The GenBank accession number from NCBI or the information of voucher specimen from JGI, along with the corresponding references (if any), are provided after the species names. The following sequences were used: Aureoboletus raphanaceus OQ674793 (Mu et al. Citation2024), Baorangia bicolor MW308599 (Li et al. Citation2021), Boletus edulis BED1_JGI (unpublished), Boletus edulis MW308609 (Li et al. Citation2021), Boletus sp. MW308606 (Li et al. Citation2021), Boletus sp. MW308608 (Li et al. Citation2021), Boletus subvelutipes MW308604 (Li et al. Citation2021), Butyriboletus hainanensis OR438787 (Zeng et al. Citation2024), Butyriboletus roseoflavus MW308600, MZ202357 (Li et al. Citation2021), Caloboletus calopus MW308602 (Li et al. Citation2021), Gyrodon lividus BX_JGI (unpublished), Hortiboletus coccyginus 2016PMI039_JGI (unpublished), Imleria badia 8406_JGI (unpublished), Lanmaoa macrocarpa OR004349 (Zheng et al. Citation2023), Neoboletus brunneissimus MW308605 (Li et al. Citation2021), Neoboletus magnificus MW308603 (Li et al. Citation2021), Neoboletus obscureumbrinus MW308607 (Li et al. Citation2021), Paxillus involutus MK993563 (Li et al. Citation2021), Paxillus rubicundulus MK993564 (Li et al. Citation2021), Pulveroboletus ravenelii NC_061666 (Cho et al. Citation2022), Retiboletus ornatipes MW308601 (Li et al. Citation2021), and Tylopilus plumbeoviolaceoides MW660363 (Li et al. Citation2021). The newly sequenced mitogenome is marked in red. Numbers near the nodes indicate bootstrap support values (>50%) and posterior probabilities (>0.95). The scale bar refers to 0.1 nucleotide substitutions per character.
Supplemental Material
Download TIFF Image (31.2 MB)Data availability statement
The mitochondrial genome data are available with the accession number of OR619662 in the GenBank of NCBI (https://www.ncbi.nlm.nih.gov/). And the associated BioProject, SRA, and BioSample numbers are PRJNA957944, SRS17371855, and SAMN34274286, respectively.
