Figures & data
Figure 1. Species reference photograph. (A) Colony morphology of Clonostachys farinosa strain CSC22A0184 on potato dextrose agar incubated at 25 °C in 7 days. The scale bar indicates 1 cm. (B) Lichen Parmotrema clavuliferum as an isolation source. The scale bar indicates 1 cm. Photograph by Seung-Yoon Oh.
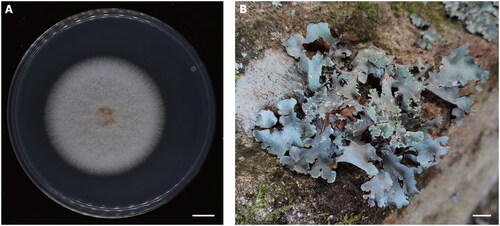
Figure 2. Circular maps and genetic compositions of Clonostachys farinosa strain CSC22A0184. Genes are located outside and color-coded by the functional classification. GC contents are shown on the inner circle. The names of genes containing introns are marked with an asterisk, and the names of genes or ORFs assigned inside the intron are located inside the outer circle.
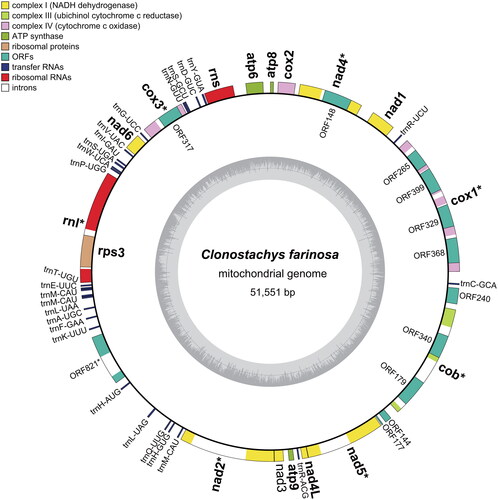
Figure 3. Maximum likelihood (ML) tree for 10 species within the order hypocreales based on 14 concatenated, conserved protein-coding genes (atp6, atp8, atp9, cob, cox1–3, nad1–6, and nad4L). The ML tree was constructed using the PROTGAMMA + MTZOA model and 1000 bootstrap replicates. The scale bar indicates the number of substitutions per site. The following mitogenomes were used for the analysis: Clonostachys rosea (Wang et al. Citation2017), C. compactiuscula, C. rogersoniana, C. solani, Clonostachys sp. YFCC 8591 (Zhao et al. Citation2021), Hapsidospora chrysogena (Eldarov et al. Citation2015), Emericellopsis fuci (Konovalova and Logacheva Citation2016), Trichoderma harzianum (Kwak Citation2021) and T. lixii (Venice et al. Citation2020). Accession numbers of GenBank are indicated after the species name.
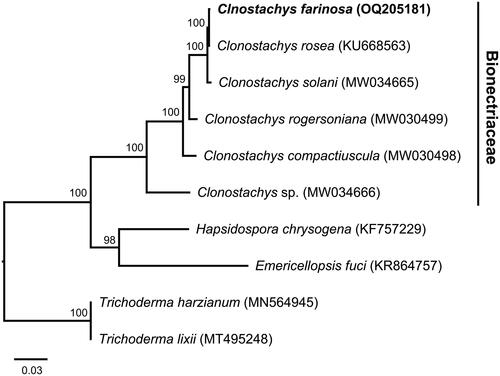
Figure 4. The collinearity analysis of mitochondrial genomes of Clonostachys species. The diagram is generated using genoPlotR package (Guy et al. Citation2010) based on the mitochondrial genome used in the phylogenomic analysis. The color represents the type of gene features.
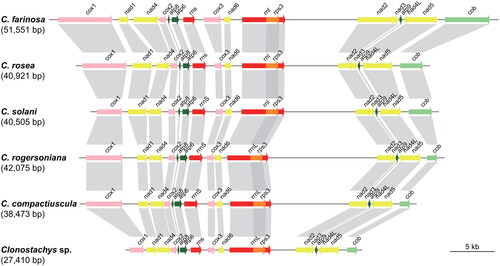
Table 1. Classification of intron and ORF annotated in the mitochondrial genome of Clonostachys farinosa.
Supplemental Material
Download PDF (295.7 KB)Data availability statement
The sequence generated from this study has been deposited to GenBank under accession number OQ205181. The associated BioProject, BioSample, and SRA numbers are PRJNA1092717, SAMN40632650, and SRR28478378, respectively.
