Figures & data
Figure 1. Reference image of the C. brevifolius used in this study. This image was taken by the authors (Xia Fan). The most characteristic feature of the specimen: leaves shorter to slightly longer than culm, mouth obliquely truncate, apex acuminate; spike single, rarely 2 or 3, globose to ovoid-globose, with numerous densely arranged spikelets.

Figure 2. Genome map of C. brevifolius chloroplast genome was generated using CPGview. The map contains six tracks. From the inner circle, the first track depicts the dispersed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The second track shows the long tandem repeats as short blue bars. The third track displays the short tandem repeats or microsatellite sequences as short bars with different colors. The fourth track depicts the sizes of the inverted repeats (IRa and IRb), small single-copy (SSC), and large single-copy (LSC). The fifth track plots the distribution of GC contents along the plastome. The sixth track displays the genes belonging to different functional groups with different colored boxes. The outer and inner genes are transcribed in the clockwise and counterclockwise directions, respectively.
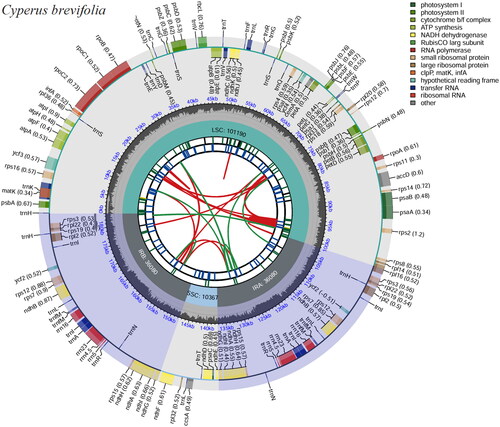
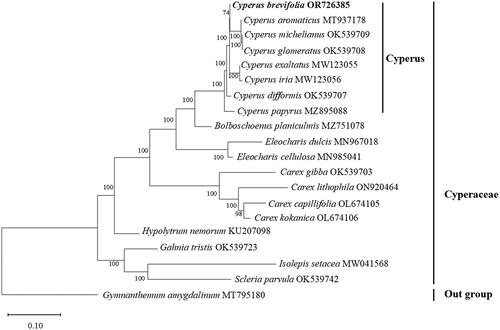
Figure 3. Maximum-likelihood phylogenetic tree of seven related species belonging to Cyperus and the placement of C. brevifolius, 11 species belonging to seven genus of Cyperaceae. Gymnanthemum amygdalinum (Asteraceae) was used as an outgroup. The bootstrap support values are shown on the nodes. The following sequences were used: Cyperus brevifolius OR726385 (this study), Cyperus aromaticus MT937178, Cyperus exaltatus MW123055, Cyperus iria MW123056 (Yang et al. Citation2021), Cyperus difformis OK539707 (Wu et al. Citation2021), Cyperus papyrus MZ895088, Cyperus michelianus OK539709 (Wu et al. Citation2021), Cyperus glomeratus OK539708 (Wu et al. Citation2021), Bolboschoenus planiculmis MZ751078 (Ning et al. Citation2024), Eleocharis dulcis MN967018 (Lee et al. Citation2020), Eleocharis cellulosa MN985041 (Lee et al. Citation2020), Carex gibba OK539703 (Wu et al. Citation2021), Carex lithophila ON920464 (Xu et al. Citation2023), Carex capillifolia OL674105, Carex kokanica OL674106, Hypolytrum nemorum KU207098, Gahnia tristis OK539723 (Wu et al. Citation2021), Isolepis setacea MW041568, Scleria parvula OK539742 (Wu et al. Citation2021), and Gymnanthemum amygdalinum MT795180 (Zhou et al. Citation2021).
Supplemental Material
Download MS Word (80.8 MB)Supplemental Material
Download TIFF Image (44.4 MB)Supplemental Material
Download TIFF Image (69.7 MB)Supplemental Material
Download TIFF Image (9.8 MB)Supplemental Material
Download MS Word (13.3 KB)Supplemental Material
Download MS Word (12.3 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of the NCBI under the accession number of OR726385 (https://www.ncbi.nlm.nih.gov/nuccore/OR726385.1/). The associated BioProject, Bio-Sample, and SRA numbers are PRJNA1060347, SAMN39225578, and SRR27427975, respectively.
