Figures & data
Figure 1. Meroplius fukuharai (Iwasa). ♂ body in left lateral view. The photo was taken by Yingying Song on 7 September 2023.

Figure 2. Gene map of the mitochondrial genome of Meroplius fukuharai (GenBank accession number: OR270120), with 13 protein-coding genes, 22 tRNAs, and two rRNAs.
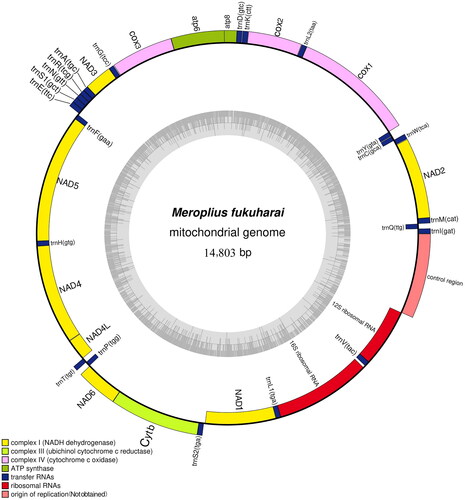
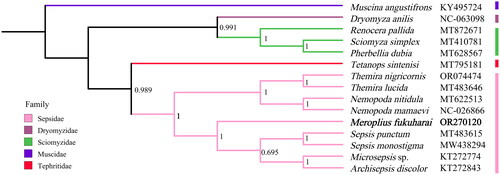
Figure 3. Bayesian phylogenetic tree of 15 Diptera species. The posterior probabilities are labeled at each node (0.6 and greater). GenBank accession numbers of all sequence used in the phylogenetic tree have been included in figure and corresponding to the names of all species. The following sequences were used: Archisepsis discolor (KT272843, Junqueira et al. Citation2016), Dryomyza anilis (NC_063098), Meroplius fukuharai (OR270120), Microsepsis sp. (KT272774, Junqueira et al. Citation2016), Muscina angustifrons (KY495724, Karagozlu et al. Citation2017), Nemopoda mamaevi (NC_026866, Li et al. Citation2015), Nemopoda nitidula (MT622513), Pherbellia dubia (MT628567), Renocera pallida (MT872671), Sciomyza simplex (MT410781), Sepsis monostigma (MW438294, Yang and Wang Citation2021), Sepsis punctum (MT483615), Tetanops sintenisi (MT795181), Themira lucida (MT483646), and Themira nigricornis (OR074474, Wong et al. Citation2023).
Supplemental Material
Download PNG Image (238.1 KB)Supplemental Material
Download PNG Image (15.2 KB)Supplemental Material
Download PNG Image (250.6 KB)Supplemental Material
Download MS Word (525.1 KB)Supplemental Material
Download MS Word (15.2 KB)Supplemental Material
Download MS Word (14.1 KB)Supplemental Material
Download PNG Image (90.4 KB)Supplemental Material
Download PDF (168.7 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OR270120.
