Figures & data
Figure 1. Circular map of the complete mitochondrial genome of Synuchus nitidus. The total length of the complete mitochondrial genome is 16,392 bp. The two outer circular bands indicate the strand and order of the genes, of which the exterior ones are on the heavy strand and the interior ones are on the light strand. The inner circle indicates the GC-skew, which is the deviation from the average GC content of the entire sequence. The photograph of S. nitidus was taken by DK.
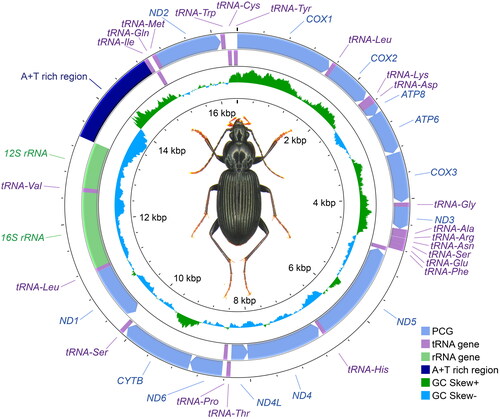
Figure 2. Inferred maximum likelihood tree based on nucleotide sequences of 13 PCGs of 36 species. As for included taxa, the family Dytiscidae was used as an outgroup. The star mark indicates the species studied here. Numbers on the branch indicate maximum likelihood support values. The following sequences were used: Hydroporus planus MW465248 (Villastrigo et al. Citation2021), Trachypachus holmbergi EU877954 (Sheffield et al. Citation2008), Cicindela anchoralis MG253029 (Wang et al. Citation2018), Manticora tibialis MF497821 (López-López and Vogler Citation2017), Pogonostoma subtiligrossum MF497820 (López-López and Vogler Citation2017), Omus cazieri MF497813 (López-López and Vogler Citation2017), Carabus changeonleei MG253028 (Wang et al. Citation2019), Carabus lafossei KY992943 (Liu et al. Citation2018), Carabus mirabilissimus GQ344500 (Wan et al. Citation2012), Carabus smaragdinus MN480425 (Oh et al. Citation2019), Harpalus pennsylvaticus MN245975 (Kieran Citation2020), Harpalus sinicus MN310888 (Yu et al. Citation2019), Abax parallelepipedus KT876877 (Linard et al. Citation2016), Pterostichus madidus KT876910 (Linard et al. Citation2016), Nebria brevicollis KT876906 (Linard et al. Citation2016), Amara aulica MN335930 (Li et al. Citation2020), Notiophilus quadripunctatus MW800883 (Raupach et al. Citation2022), Omophron limbatum MW800882 (Raupach et al. Citation2022), Metrius contractus MF497817 (López-López and Vogler Citation2017), Scarites buparius MF497822 (López-López and Vogler Citation2017), Brachinus crepitans JX412826, Carabus hortensis MN122850, Carabus granulatus MN122870, Blethisa multipunctata KX087243, Elaphrus cupreus KX087286, Hexagonia terminalis JX412768, Craspedophorus nobilis JX412738, Pterostichus niger KX087231, Pterostichus oblongopunctatus MN122833, Stomis pumicatus KX087349, Amara communis KX035135, Lebia chlorocephala KX087304, Promecognathus crassus JX313665, Bembidion varium KX087242, Pogonus iridipennis KX087338, and Synuchus nitidus OR755976.

Supplemental Material
Download MS Word (231.2 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank at http://www.ncbi.nlm.nih.gov/, under the accession number OR755976. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1051788, SRX22874416, and SAMN38797855, respectively.
