Figures & data
Figure 1. Devario shanensis from the Nangun River in Cangyuan Prefecture, Yunnan province, China (23.2254° N, 98.9899° E). (photo by Xiao Jiang CHEN on 20 July 2022).

Table 1. Species names, corresponding GenBank accession numbers, and references for all 14 mitogenomes utilized in the reconstruction of the phylogenetic tree.
Figure 2. The mitochondrial genome gene map of devario shanensis displays the localization of genes situated on the heavy and light strands, with arrows pointing in opposite directions to indicate their transcriptional orientation. Genes encoded on the heavy strand are depicted outside the circle, while those on the light strand are shown inside.
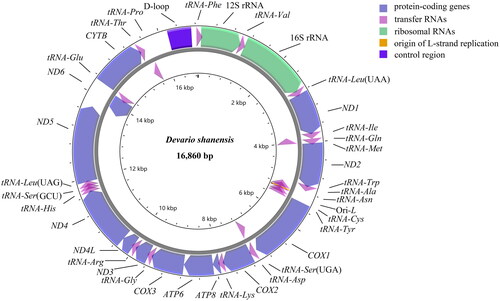
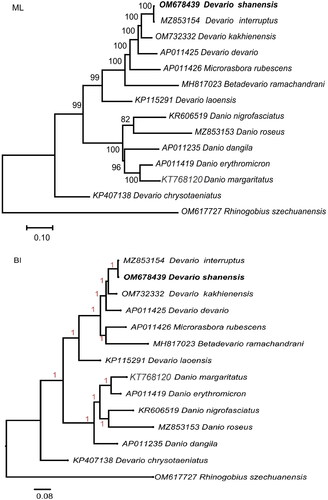
Figure 3. Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees were constructed using complete mitochondrial genomes of Devario shanensis and 13 other fish species. The following sequences were used: MZ853154 (Chen et al. Citation2023), OM732332 (Song et al. Citation2022), AP011425 (Tang et al. Citation2010), AP011426 (Tang et al. Citation2010), MH817023 (Norén & kullander Citation2018), KP115291 (Zhang et al. 2016), KR606519 (Huang et al. Citation2016), MZ853153 (Song et al. Citation2022), AP011235 (Tang et al. Citation2010), AP011419(Tang et al. Citation2010), KT768120 (unpublished), KP407138 (Liu et al. Citation2016), rhinogobius szechuanensis OM617727 (Liu et al. Citation2023). ML bootstrap support values and Bayesian posterior probabilities were depicted above branches. Each species was accompanied by its GenBank accession number.
Supplemental Material
Download MS Word (167.9 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM678439.
The associated "BioProject", "Bio-Sample" and "SRA" numbers are PRJNA808163, SAMN26029380, and SRR18063683 respectively.
