Figures & data
Figure 1. Species reference image of B. scoparia. The photos were taken by Miss Lin Li at the germplasm resource nursery of Enshi Tujia and Miao Autonomous Prefecture Academy of Agricultural Sciences, Hubei Province, China, in September 2023.

Figure 2. The map of B. scoparia chloroplast genome by CPGview. The map consists of six tracks. From the center to the outer, the first track shows dispersed repeats connected by red and green arcs indicating the direction (forward and reverse, respectively). The second track shows long tandem repeats as blue bands, and the third track shows short tandem repeats or microsatellites as green bands. The fourth track represents the GC content along the plastome. Finally, the sixth track represents the genes as colored boxes, the inner boxes present clockwise transcription, and the outer boxes present counterclockwise transcribed genes.
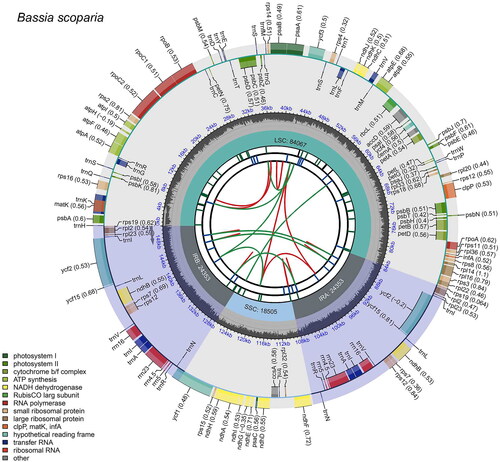
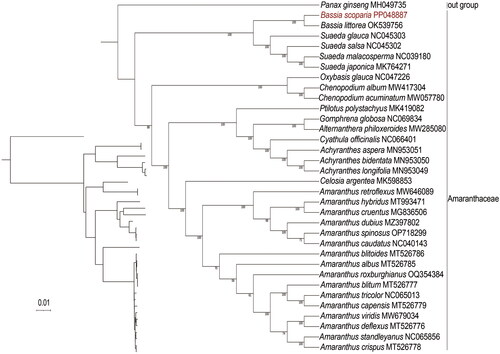
Figure 3. Phylogenetic relationships of B. scoparia based on the maximum-likelihood (ML) analysis of protein-coding genes in chloroplast genomes. Bootstrap values next to the nodes are based on 1000 replications. Panax ginseng was set as the outgroup. GenBank accession numbers: Panax ginseng MH049735 (Wang et al. Citation2018), Bassia littorea OK539756, Suaeda glauca NC045303 (Qu et al. Citation2019a), Suaeda salsa NC045302 (Qu et al. Citation2019b), Suaeda malacosperma NC039180 (Park et al. Citation2018), Suaeda japonica MK764271 (Park et al. Citation2018), Oxybasis glauca NC047226, Chenopodium album MW417304 (Li et al. Citation2021), Chenopodium acuminatum MW057780 (Wariss and Qu Citation2021), Celosia argentea MK598853 (Qian et al. Citation2019), Ptilotus polystachyus MK419082 (Hammer et al. Citation2019), Gomphrena globosa NC069834, Alternanthera philoxeroides MW285080 (Jiang et al. Citation2021), Cyathula officinalis NC066401 (Guo et al. Citation2022), Achyranthes aspera MN953051 (Xu et al. Citation2020), Achyranthes bidentata MN953050 (Xu et al. Citation2020), Achyranthes longifolia MN953049 (Xu et al. Citation2020), Celosia argentea MK598853(Qian et al. Citation2019), Amaranthus retroflexus MW646089 (Lou and Fan Citation2021), Amaranthus hybridus MT993471 (Bai et al. Citation2021), Amaranthus cruentus MG836506 (Hong et al. Citation2019), Amaranthus dubius MZ397802 (Xu et al. Citation2021), Amaranthus spinosus OP718299, Amaranthus caudatus NC040143 (Hong et al. Citation2019), Amaranthus blitoides MT526786 (Xu et al. Citation2022), Amaranthus albus MT526785 (Xu et al. Citation2022), Amaranthus roxburghianus OQ354384, Amaranthus blitum MT526777 (Xu et al. Citation2022), Amaranthus tricolor NC065013, Amaranthus capensis MT526779 (Xu et al. Citation2022), Amaranthus viridis MW679034 (Ding et al. Citation2021), Amaranthus deflexus MT526776 (Xu et al. Citation2022), Amaranthus standleyanus NC065856 (Xu et al. Citation2022), and Amaranthus crispus MT526778 (Xu et al. Citation2022).
Supplemental Material
Download MS Word (544.1 KB)Supplemental Material
Download MS Excel (13.7 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. PP048887. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1063496, SRR27534540, and SAMN39397596, respectively.
