Figures & data
Figure 1. Morphological characteristics of Olea dioica. The photo was taken by the author Zhaoshan Wang at the Research Institute of Forestry, Chinese Academy of Forestry. The leaves of O. dioica are blade leathery, lanceolate, oblanceolate or long elliptic-lanceolate, apex acuminate or obtuse, sparsely rounded, base cuneate, entire or irregularly and sparsely serrate, usually reddish when dry.

Figure 2. The circular map of the chloroplast genome of Olea dioica. Genes belonging to different functional groups are plotted in the outer circle. The functional classification of the genes is shown at the bottom left. The dark gray in the inner circle indicates the GC content of the chloroplast genome. The quadripartite structure, which consists of the LSC, the SSC, and two IR regions, is shown. The inner track shows the forward and reverse repeats connected with red and green arcs.
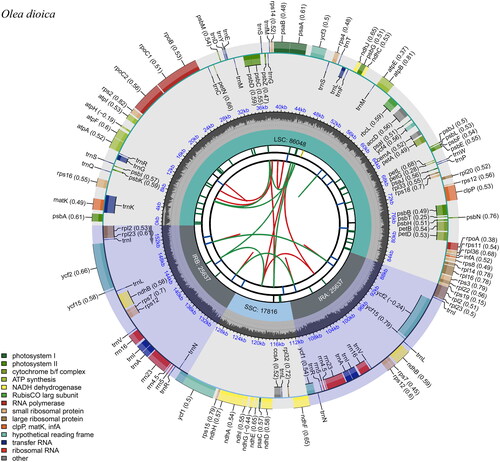
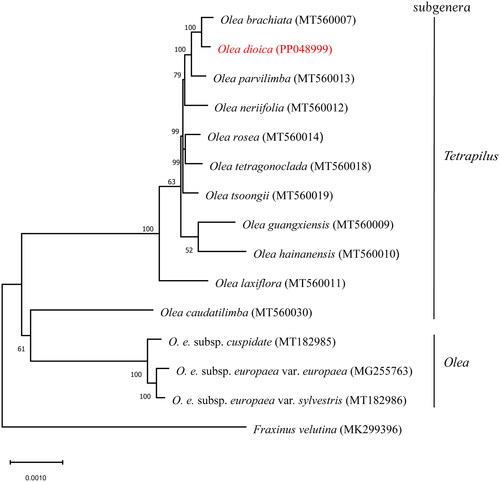
Figure 3. Maximum-likelihood (ML) phylogenetic tree with evolutionary distances for Olea dioica and 14 related taxa from the family Oleaceae. Fraxinus velutina is used as an outgroup. Numbers at the nodes indicated bootstrap support values from 1000 replicates. Accession numbers: Olea brachiata, MT560007 (Dong et al. Citation2022); Olea dioica (this study); Olea parvilimba, MT560013 (Dong et al. Citation2022); Olea neriifolia, MT560012 (Dong et al. Citation2022); Olea rosea, MT560014 (Dong et al. Citation2022); Olea tetragonoclada, MT560018 (Dong et al. Citation2022); Olea tsoongii, MT560019 (Dong et al. Citation2022); Olea guangxiensis, MT560009 (Dong et al. Citation2022); Olea hainanensis, MT560010 (Dong et al. Citation2022); Olea laxiflora, MT560011 (Dong et al. Citation2022); Olea europaea subsp. cuspidata, MT182985 (Niu et al. Citation2020); Olea europaea subsp. europaea var. europaea, MG255763 (reference unavailable); Olea europaea subsp. europaea var. sylvestris, MT182986 (Niu et al. Citation2020); Olea caudatilimba, MT560030 (Dong et al. Citation2022); Fraxinus velutina, MK299396 (Olofsson et al. Citation2019).
Supplemental Material
Download TIFF Image (356.1 KB)Supplemental Material
Download TIFF Image (755.6 KB)Supplemental Material
Download TIFF Image (1.1 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number PP048999. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1061655, SRR27469441, and SAMN39276719, respectively.
